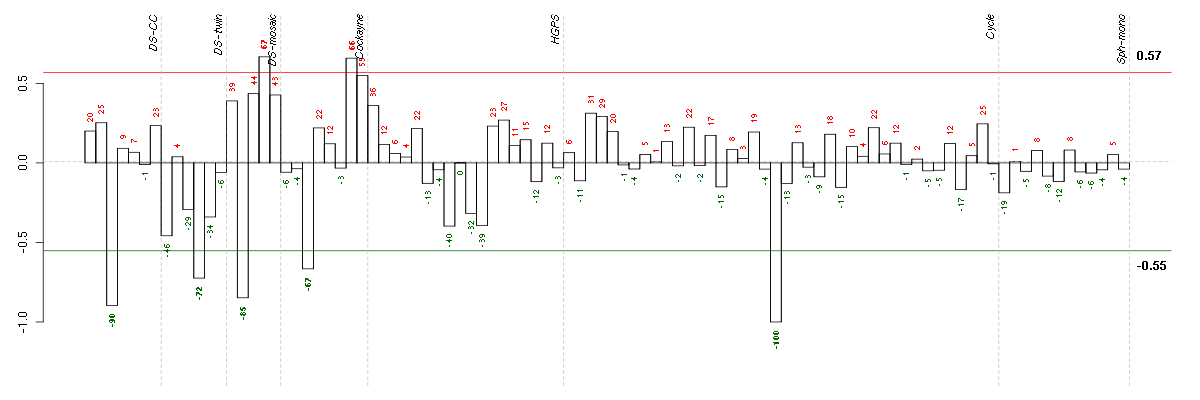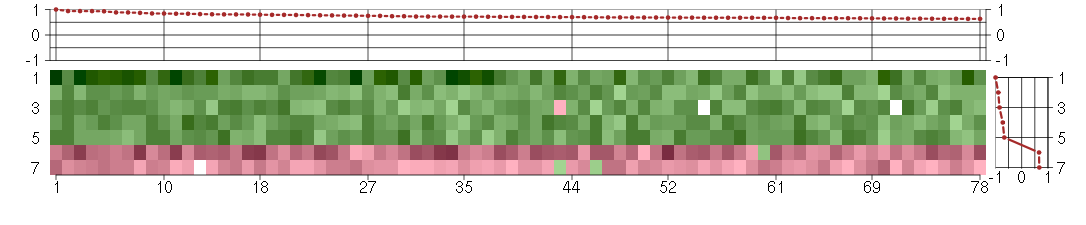

Under-expression is coded with green,
over-expression with red color.

response to mechanical stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a mechanical stimulus.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
response to abiotic stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an abiotic (non-living) stimulus.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular component organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of a cellular component.
membrane organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of a membrane. A membrane is a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
organelle localization
Any process by which an organelle is transported to, and/or maintained in, a specific location.
cellular localization
Any process by which a substance or cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in, a specific location within or in the membrane of a cell.
all
This term is the most general term possible
cellular localization
Any process by which a substance or cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in, a specific location within or in the membrane of a cell.
response to mechanical stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a mechanical stimulus.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
membrane-bounded vesicle
Any small, fluid-filled, spherical organelle enclosed by a lipid bilayer.
melanosome
A tissue-specific, membrane-bounded cytoplasmic organelle within which melanin pigments are synthesized and stored. Melanosomes are synthesized in melanocyte cells.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
pigment granule
A small, subcellular membrane-bounded vesicle containing pigment and/or pigment precursor molecules. Pigment granule biogenesis is poorly understood, as pigment granules are derived from multiple sources including the endoplasmic reticulum, coated vesicles, lysosomes, and endosomes.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.

ADAM15ADAM metallopeptidase domain 15 (217007_s_at), score: 0.69 ADRBK1adrenergic, beta, receptor kinase 1 (201401_s_at), score: 0.66 AP1S1adaptor-related protein complex 1, sigma 1 subunit (205195_at), score: 0.66 ARF1ADP-ribosylation factor 1 (208750_s_at), score: 0.71 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 0.79 ATP13A3ATPase type 13A3 (219558_at), score: 0.72 ATP6V1AATPase, H+ transporting, lysosomal 70kDa, V1 subunit A (201971_s_at), score: 0.72 BCL2L1BCL2-like 1 (215037_s_at), score: 0.68 C19orf6chromosome 19 open reading frame 6 (213986_s_at), score: 1 CALUcalumenin (214845_s_at), score: 0.69 CANXcalnexin (208853_s_at), score: 0.64 CD44CD44 molecule (Indian blood group) (210916_s_at), score: 0.82 CDV3CDV3 homolog (mouse) (213548_s_at), score: 0.78 CLIC4chloride intracellular channel 4 (201559_s_at), score: 0.65 COL1A1collagen, type I, alpha 1 (217430_x_at), score: 0.67 COL6A1collagen, type VI, alpha 1 (212940_at), score: 0.93 COPAcoatomer protein complex, subunit alpha (214336_s_at), score: 0.94 CPDcarboxypeptidase D (201942_s_at), score: 0.77 DAPK3death-associated protein kinase 3 (203890_s_at), score: 0.81 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: 0.83 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: 0.8 EIF5Aeukaryotic translation initiation factor 5A (201123_s_at), score: 0.68 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: 0.76 ERLIN1ER lipid raft associated 1 (202444_s_at), score: 0.63 FGFR1fibroblast growth factor receptor 1 (210973_s_at), score: 0.75 FN1fibronectin 1 (214701_s_at), score: 0.88 GLULglutamate-ammonia ligase (glutamine synthetase) (200648_s_at), score: 0.64 GNSglucosamine (N-acetyl)-6-sulfatase (203676_at), score: 0.68 GPR137G protein-coupled receptor 137 (43934_at), score: 0.69 GTF2Igeneral transcription factor II, i (210892_s_at), score: 0.72 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.71 HP1BP3heterochromatin protein 1, binding protein 3 (220633_s_at), score: 0.71 KCTD20potassium channel tetramerisation domain containing 20 (214849_at), score: 0.72 LAMP1lysosomal-associated membrane protein 1 (201551_s_at), score: 0.84 LOC285412similar to KIAA0737 protein (217448_s_at), score: 0.66 LOXL2lysyl oxidase-like 2 (202997_s_at), score: 0.7 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (211019_s_at), score: 0.65 MAPKAPK2mitogen-activated protein kinase-activated protein kinase 2 (201461_s_at), score: 0.68 MAT2Amethionine adenosyltransferase II, alpha (200769_s_at), score: 0.72 MAXMYC associated factor X (210734_x_at), score: 0.84 MFN2mitofusin 2 (216205_s_at), score: 0.66 MMP14matrix metallopeptidase 14 (membrane-inserted) (202827_s_at), score: 0.69 MPDU1mannose-P-dolichol utilization defect 1 (209208_at), score: 0.64 NUMA1nuclear mitotic apparatus protein 1 (214251_s_at), score: 0.68 PAFAH1B1platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa (211547_s_at), score: 0.71 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.89 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.94 PHTF2putative homeodomain transcription factor 2 (217097_s_at), score: 0.72 PICALMphosphatidylinositol binding clathrin assembly protein (215236_s_at), score: 0.74 PIP5K1Aphosphatidylinositol-4-phosphate 5-kinase, type I, alpha (211205_x_at), score: 0.71 PLEC1plectin 1, intermediate filament binding protein 500kDa (216971_s_at), score: 0.67 POLDIP3polymerase (DNA-directed), delta interacting protein 3 (215357_s_at), score: 0.76 PRKAB1protein kinase, AMP-activated, beta 1 non-catalytic subunit (201835_s_at), score: 0.63 PRPF6PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) (208879_x_at), score: 0.66 PRRX1paired related homeobox 1 (205991_s_at), score: 0.83 PTBP1polypyrimidine tract binding protein 1 (212016_s_at), score: 0.94 PTPN14protein tyrosine phosphatase, non-receptor type 14 (205503_at), score: 0.75 PXNpaxillin (211823_s_at), score: 0.81 RAB35RAB35, member RAS oncogene family (205461_at), score: 0.74 RAB5ARAB5A, member RAS oncogene family (206113_s_at), score: 0.81 RAB5CRAB5C, member RAS oncogene family (201156_s_at), score: 0.68 RCAN1regulator of calcineurin 1 (215253_s_at), score: 0.69 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: 0.64 RXRBretinoid X receptor, beta (215099_s_at), score: 0.77 SEPT11septin 11 (201308_s_at), score: 0.7 SLC12A4solute carrier family 12 (potassium/chloride transporters), member 4 (209401_s_at), score: 0.8 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (207043_s_at), score: 0.65 SNAP23synaptosomal-associated protein, 23kDa (214544_s_at), score: 0.68 SPPL2Bsignal peptide peptidase-like 2B (215833_s_at), score: 0.64 SRPRsignal recognition particle receptor (docking protein) (200917_s_at), score: 0.79 STIP1stress-induced-phosphoprotein 1 (212009_s_at), score: 0.78 SYPL1synaptophysin-like 1 (201259_s_at), score: 0.7 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.87 TMED2transmembrane emp24 domain trafficking protein 2 (204426_at), score: 0.66 WACWW domain containing adaptor with coiled-coil (219679_s_at), score: 0.71 WDR1WD repeat domain 1 (210935_s_at), score: 0.69 YIPF5Yip1 domain family, member 5 (221423_s_at), score: 0.66 YWHAEtyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide (210317_s_at), score: 0.68
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |