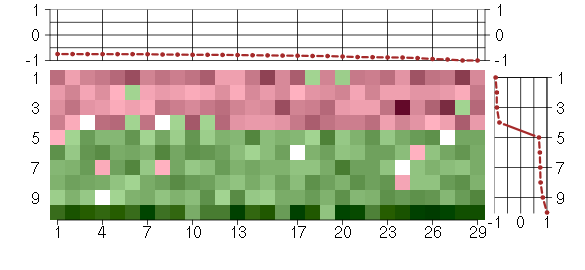

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
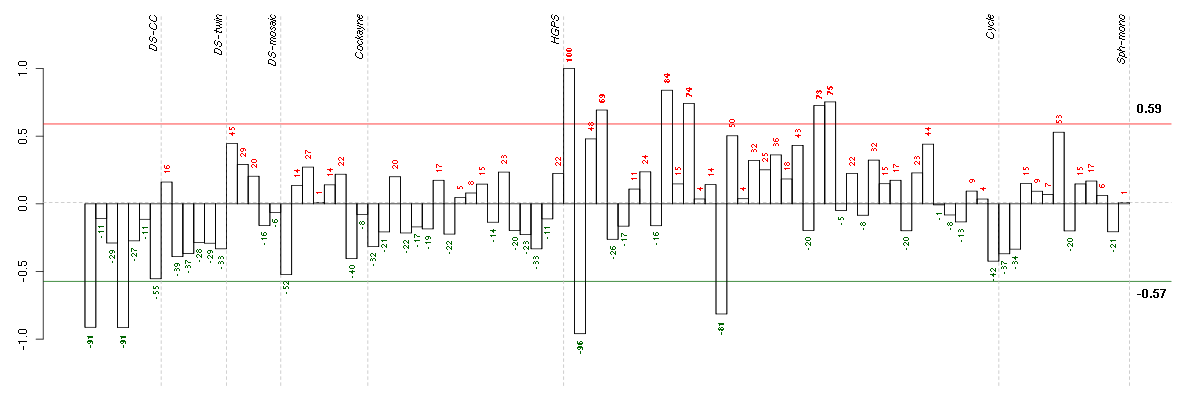
ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: -0.8 ADORA1adenosine A1 receptor (216220_s_at), score: -0.87 ADRA1Dadrenergic, alpha-1D-, receptor (210961_s_at), score: -0.75 AZGP1alpha-2-glycoprotein 1, zinc-binding (217014_s_at), score: -0.77 CATSPER2cation channel, sperm associated 2 (217588_at), score: -0.77 CCDC9coiled-coil domain containing 9 (206257_at), score: -0.88 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: -0.79 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.84 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: -0.75 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: -1 HAB1B1 for mucin (215778_x_at), score: -0.83 LILRA5leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 (215838_at), score: -0.78 LOC80054hypothetical LOC80054 (220465_at), score: -0.81 LRRN2leucine rich repeat neuronal 2 (216164_at), score: -0.81 LTBP4latent transforming growth factor beta binding protein 4 (210628_x_at), score: -0.78 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: -0.75 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: -0.91 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: -0.94 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.79 NOTCH4Notch homolog 4 (Drosophila) (205247_at), score: -0.75 PIPOXpipecolic acid oxidase (221605_s_at), score: -0.78 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: -0.96 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: -0.82 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: -0.75 SLC26A10solute carrier family 26, member 10 (214951_at), score: -0.75 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: -0.75 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.87 TRA@T cell receptor alpha locus (216540_at), score: -1 VGFVGF nerve growth factor inducible (205586_x_at), score: -0.88
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486111.cel | 24 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |