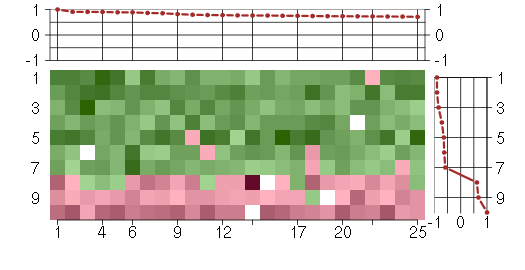

Under-expression is coded with green,
over-expression with red color.



CD53CD53 molecule (203416_at), score: 0.78 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.72 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: 0.89 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.91 FBRSfibrosin (218255_s_at), score: 0.74 FBXO2F-box protein 2 (219305_x_at), score: 0.71 FGL1fibrinogen-like 1 (205305_at), score: 0.86 GPR144G protein-coupled receptor 144 (216289_at), score: 0.91 HAB1B1 for mucin (215778_x_at), score: 1 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.89 ITPKCinositol 1,4,5-trisphosphate 3-kinase C (213076_at), score: 0.76 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: 0.72 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.8 NACA2nascent polypeptide-associated complex alpha subunit 2 (222224_at), score: 0.76 PGCprogastricsin (pepsinogen C) (205261_at), score: 0.85 PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.78 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.9 SAA4serum amyloid A4, constitutive (207096_at), score: 0.75 SEPT5septin 5 (209767_s_at), score: 0.73 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: 0.75 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.82 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: 0.74 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.77 TNK1tyrosine kinase, non-receptor, 1 (217149_x_at), score: 0.74
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |