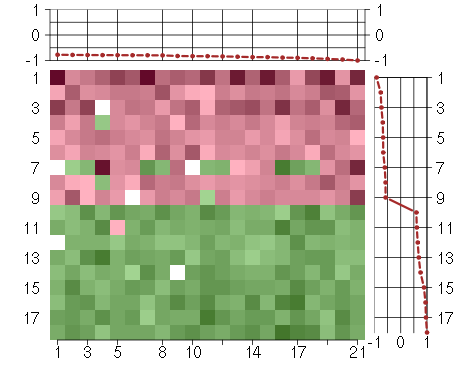

Under-expression is coded with green,
over-expression with red color.



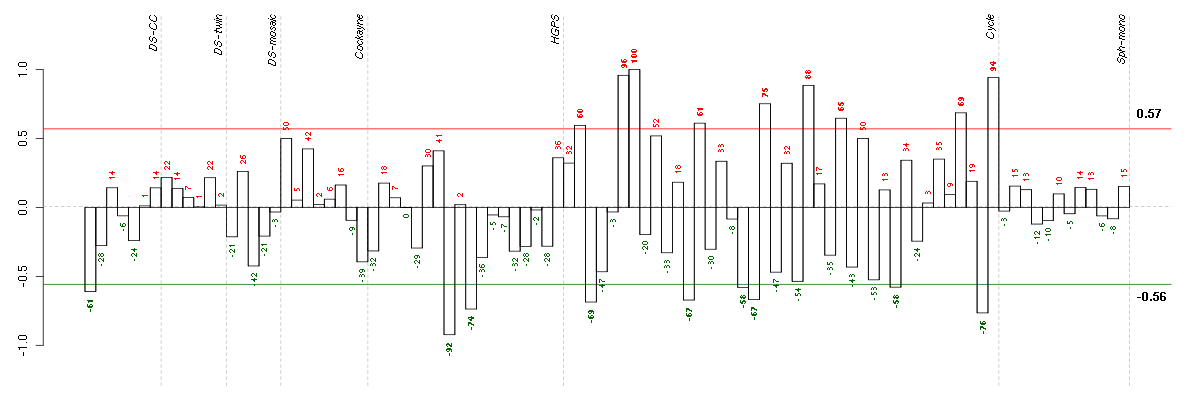
ATN1atrophin 1 (40489_at), score: -1 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.79 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.84 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.79 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: -0.83 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.89 FOXK2forkhead box K2 (203064_s_at), score: -0.9 IDUAiduronidase, alpha-L- (205059_s_at), score: -0.79 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.94 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: -0.8 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: -0.83 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: -0.85 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: -0.87 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: -0.79 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.87 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: -0.78 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.91 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.96 STRN4striatin, calmodulin binding protein 4 (217903_at), score: -0.77 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.8 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: -0.82
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |