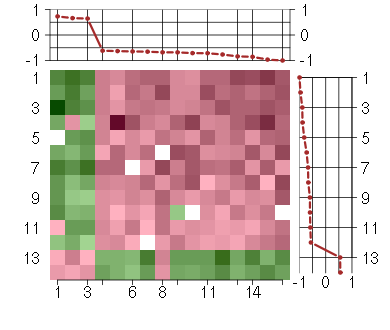

Under-expression is coded with green,
over-expression with red color.



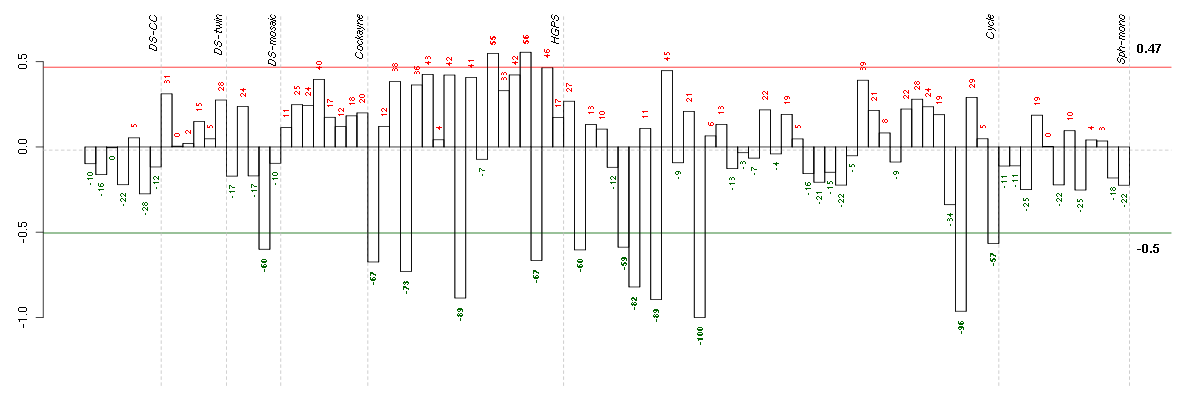
CDH13cadherin 13, H-cadherin (heart) (204726_at), score: -0.65 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.66 CUX1cut-like homeobox 1 (214743_at), score: -0.62 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.71 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.84 NEFMneurofilament, medium polypeptide (205113_at), score: -1 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.65 NRXN3neurexin 3 (205795_at), score: -0.85 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.68 PCDH9protocadherin 9 (219737_s_at), score: -0.96 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: -0.68 PRSS3protease, serine, 3 (213421_x_at), score: -0.71 SNNstannin (218032_at), score: 0.73 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: -0.77 TRY6trypsinogen C (215395_x_at), score: -0.63 UGDHUDP-glucose dehydrogenase (203343_at), score: -0.65
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690336.cel | 9 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |