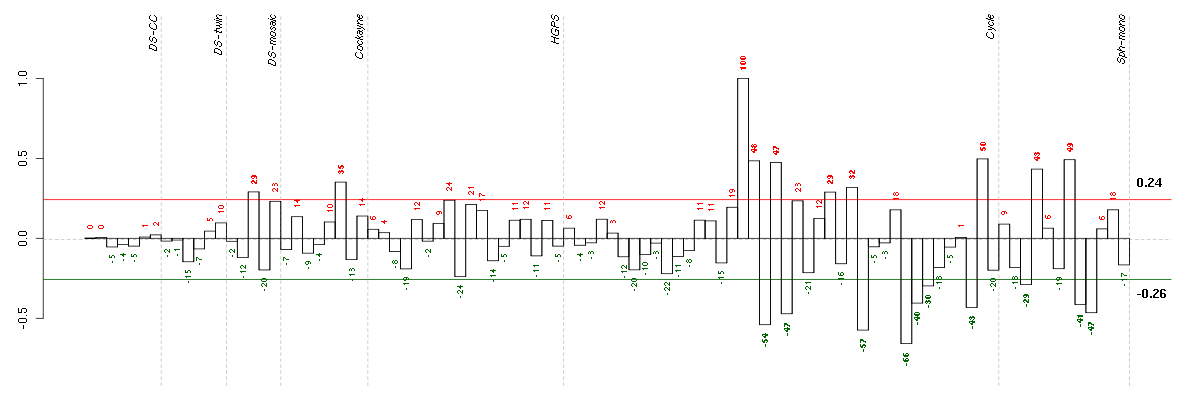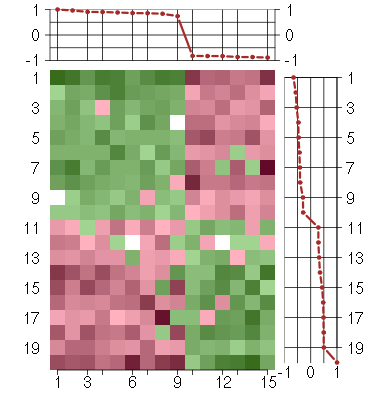

Under-expression is coded with green,
over-expression with red color.



ANKRD28ankyrin repeat domain 28 (213035_at), score: -0.83 C5orf4chromosome 5 open reading frame 4 (48031_r_at), score: -0.89 CDCA4cell division cycle associated 4 (218399_s_at), score: 0.96 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: 0.83 CYTH1cytohesin 1 (202880_s_at), score: 0.91 GIT1G protein-coupled receptor kinase interacting ArfGAP 1 (218030_at), score: 0.74 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: -0.82 IPPKinositol 1,3,4,5,6-pentakisphosphate 2-kinase (219092_s_at), score: 0.86 NEDD4Lneural precursor cell expressed, developmentally down-regulated 4-like (212445_s_at), score: 1 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: 0.88 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: -0.83 R3HDM2R3H domain containing 2 (203831_at), score: -0.86 RABL3RAB, member of RAS oncogene family-like 3 (213970_at), score: -0.87 SPAG1sperm associated antigen 1 (210117_at), score: 0.9 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: 0.85
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |