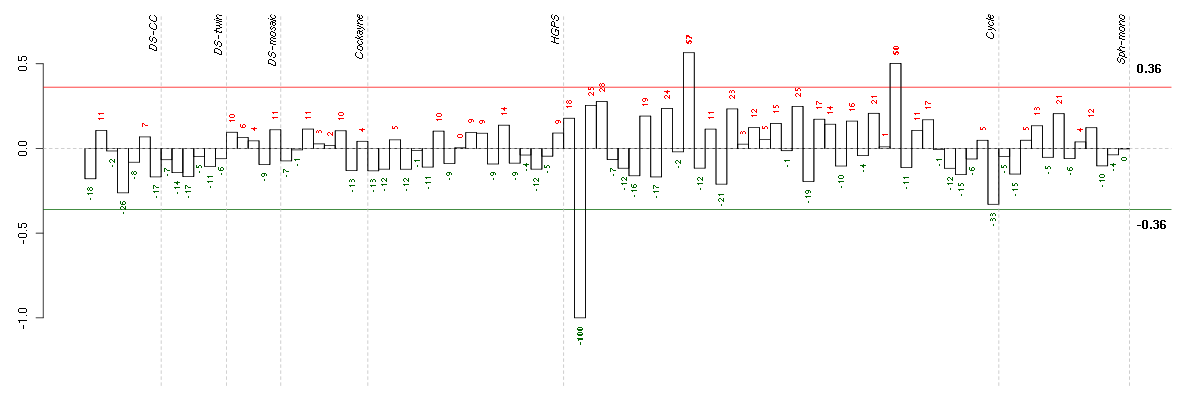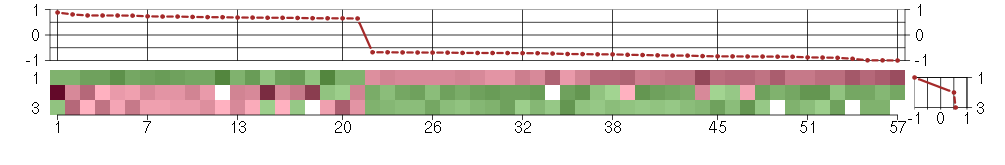

Under-expression is coded with green,
over-expression with red color.



ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: -0.83 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: -0.76 ASB1ankyrin repeat and SOCS box-containing 1 (212819_at), score: 0.68 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: -0.93 ATP8B2ATPase, class I, type 8B, member 2 (216873_s_at), score: -0.71 BCL11AB-cell CLL/lymphoma 11A (zinc finger protein) (219497_s_at), score: 0.66 BCL2L1BCL2-like 1 (215037_s_at), score: 0.65 C20orf117chromosome 20 open reading frame 117 (207711_at), score: 0.68 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: -0.85 C2orf37chromosome 2 open reading frame 37 (220172_at), score: 0.76 CA1carbonic anhydrase I (205949_at), score: 0.68 CALCAcalcitonin-related polypeptide alpha (217561_at), score: -0.68 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: -0.89 CEP164centrosomal protein 164kDa (204251_s_at), score: 0.7 CMAHcytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) pseudogene (205518_s_at), score: 0.81 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: -0.84 EMCNendomucin (219436_s_at), score: 0.65 EPS8L1EPS8-like 1 (218778_x_at), score: -0.81 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.72 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: -0.73 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: -0.84 GLRA1glycine receptor, alpha 1 (207972_at), score: -0.71 HIST1H4Ghistone cluster 1, H4g (208551_at), score: -0.78 HOOK1hook homolog 1 (Drosophila) (219976_at), score: -0.75 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: 0.66 HSPA6heat shock 70kDa protein 6 (HSP70B') (117_at), score: -0.99 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: -0.99 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: -0.84 KCTD17potassium channel tetramerisation domain containing 17 (205561_at), score: -0.84 KIF16Bkinesin family member 16B (219570_at), score: 0.76 KLHL3kelch-like 3 (Drosophila) (221221_s_at), score: -0.69 KRT8P12keratin 8 pseudogene 12 (222060_at), score: -1 LOC149501similar to keratin 8 (216821_at), score: -0.8 LOC80054hypothetical LOC80054 (220465_at), score: -0.79 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: -0.81 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: -0.7 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: -0.85 NIPSNAP3Bnipsnap homolog 3B (C. elegans) (221104_s_at), score: 0.76 OR3A2olfactory receptor, family 3, subfamily A, member 2 (221386_at), score: -0.71 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.71 PLUNCpalate, lung and nasal epithelium associated (220542_s_at), score: -0.71 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: -0.7 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.7 SH2D3ASH2 domain containing 3A (222169_x_at), score: -0.75 SORL1sortilin-related receptor, L(DLR class) A repeats-containing (203509_at), score: -0.68 SPEF1sperm flagellar 1 (216119_s_at), score: -0.69 SPTA1spectrin, alpha, erythrocytic 1 (elliptocytosis 2) (206937_at), score: 0.69 TMPRSS6transmembrane protease, serine 6 (214955_at), score: -0.76 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.68 TRIM21tripartite motif-containing 21 (204804_at), score: 0.73 TRMUtRNA 5-methylaminomethyl-2-thiouridylate methyltransferase (213634_s_at), score: -0.88 TTC30Atetratricopeptide repeat domain 30A (213679_at), score: 0.67 WFDC8WAP four-disulfide core domain 8 (215276_at), score: -0.88 ZNF318zinc finger protein 318 (203521_s_at), score: 0.72 ZNF783zinc finger family member 783 (221876_at), score: -0.69 ZSCAN16zinc finger and SCAN domain containing 16 (219676_at), score: 0.77
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |