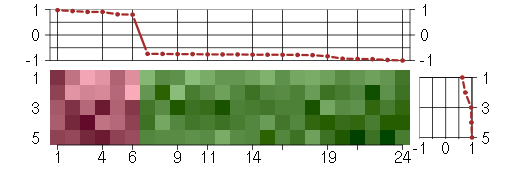

Under-expression is coded with green,
over-expression with red color.



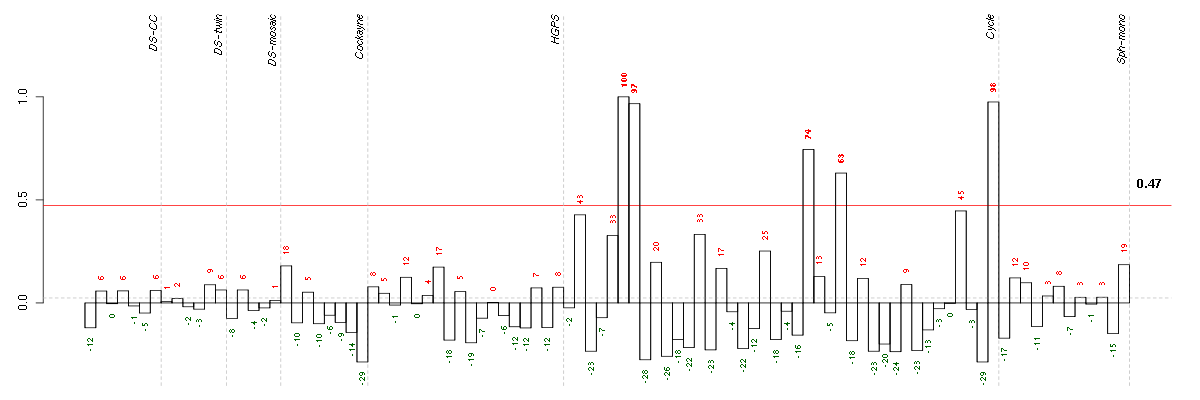
ADCY3adenylate cyclase 3 (209321_s_at), score: -0.95 CHST2carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 (203921_at), score: -0.76 COL10A1collagen, type X, alpha 1 (217428_s_at), score: -0.74 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: -0.93 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.79 FOXK2forkhead box K2 (203064_s_at), score: -1 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: -0.98 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.81 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: -0.75 MC4Rmelanocortin 4 receptor (221467_at), score: 0.94 NEFMneurofilament, medium polypeptide (205113_at), score: 0.8 NUDT11nudix (nucleoside diphosphate linked moiety X)-type motif 11 (219855_at), score: -0.76 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: -0.75 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.74 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: -0.77 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: -0.83 SNX27sorting nexin family member 27 (221006_s_at), score: -0.76 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.78 TBX3T-box 3 (219682_s_at), score: -0.78 TMEM149transmembrane protein 149 (219690_at), score: 0.97 TOP3Atopoisomerase (DNA) III alpha (214300_s_at), score: 0.91 TSKUtsukushin (218245_at), score: -0.94 WDR6WD repeat domain 6 (217734_s_at), score: -0.77 ZDHHC14zinc finger, DHHC-type containing 14 (219247_s_at), score: 0.91
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |