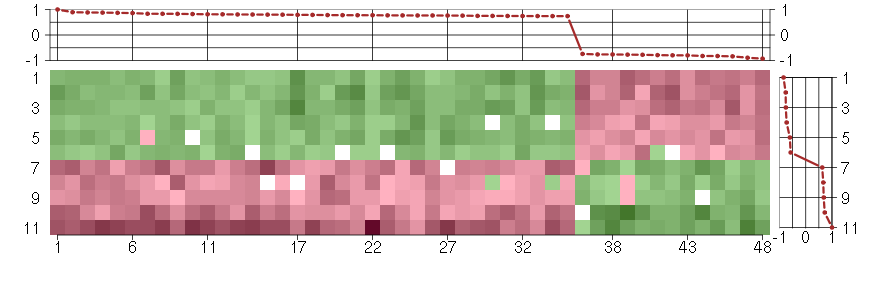

Under-expression is coded with green,
over-expression with red color.



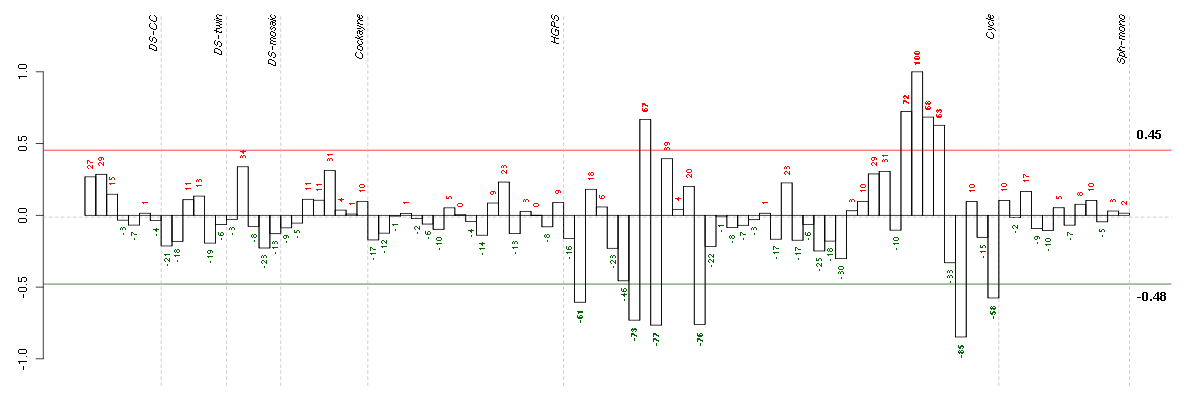
ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: 0.78 ADORA2Badenosine A2b receptor (205891_at), score: 0.78 AIM1absent in melanoma 1 (212543_at), score: 0.74 ANGPTL2angiopoietin-like 2 (213004_at), score: 0.81 ANXA10annexin A10 (210143_at), score: -0.82 APOL6apolipoprotein L, 6 (219716_at), score: 0.75 BDKRB2bradykinin receptor B2 (205870_at), score: 0.77 BTG2BTG family, member 2 (201236_s_at), score: 0.8 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: 0.89 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.79 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.79 DAPK1death-associated protein kinase 1 (203139_at), score: 0.83 DCLK1doublecortin-like kinase 1 (205399_at), score: 0.88 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.75 DEPDC6DEP domain containing 6 (218858_at), score: 0.77 DHRS3dehydrogenase/reductase (SDR family) member 3 (202481_at), score: 0.75 DOCK10dedicator of cytokinesis 10 (219279_at), score: -0.76 FGF9fibroblast growth factor 9 (glia-activating factor) (206404_at), score: 0.74 GALNT12UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) (218885_s_at), score: 0.81 GKglycerol kinase (207387_s_at), score: -0.79 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.8 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 1 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.77 IFIH1interferon induced with helicase C domain 1 (219209_at), score: 0.83 IRF1interferon regulatory factor 1 (202531_at), score: 0.78 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.74 KIAA1305KIAA1305 (220911_s_at), score: 0.76 LDLRlow density lipoprotein receptor (202068_s_at), score: 0.74 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.86 MC4Rmelanocortin 4 receptor (221467_at), score: -0.78 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.76 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: -0.83 MX2myxovirus (influenza virus) resistance 2 (mouse) (204994_at), score: 0.82 NEFMneurofilament, medium polypeptide (205113_at), score: -0.93 NPTX1neuronal pentraxin I (204684_at), score: -0.77 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.87 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: 0.81 PCDH9protocadherin 9 (219737_s_at), score: -0.84 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.74 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: 0.88 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: 0.76 SECTM1secreted and transmembrane 1 (213716_s_at), score: 0.78 SIX2SIX homeobox 2 (206510_at), score: 0.8 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.8 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.8 TLR3toll-like receptor 3 (206271_at), score: 0.83 TP53tumor protein p53 (201746_at), score: 0.77 ULBP2UL16 binding protein 2 (221291_at), score: -0.89
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |