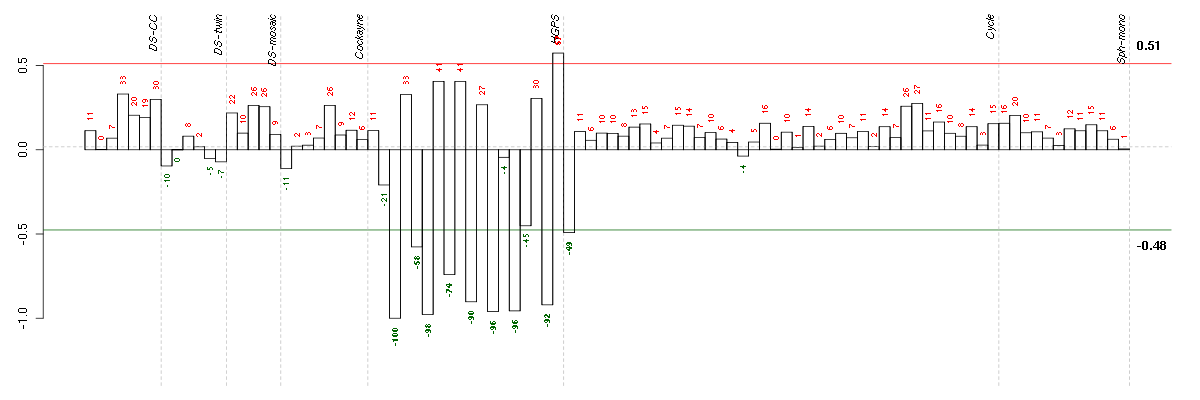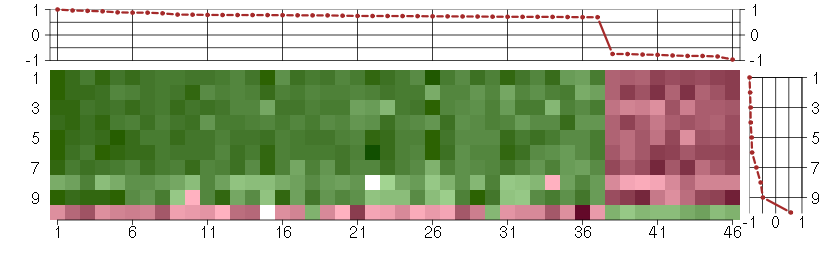

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

ANKRD1ankyrin repeat domain 1 (cardiac muscle) (206029_at), score: 0.96 AUTS2autism susceptibility candidate 2 (212599_at), score: -0.74 BST1bone marrow stromal cell antigen 1 (205715_at), score: 0.73 C1orf54chromosome 1 open reading frame 54 (219506_at), score: 0.7 C8orf84chromosome 8 open reading frame 84 (214725_at), score: 0.73 CAMK2N1calcium/calmodulin-dependent protein kinase II inhibitor 1 (218309_at), score: -0.96 CCR10chemokine (C-C motif) receptor 10 (220565_at), score: 0.72 CYP1B1cytochrome P450, family 1, subfamily B, polypeptide 1 (202436_s_at), score: 0.73 EDIL3EGF-like repeats and discoidin I-like domains 3 (207379_at), score: 0.71 EPHA5EPH receptor A5 (215664_s_at), score: 0.95 F2Rcoagulation factor II (thrombin) receptor (203989_x_at), score: 0.78 FGL2fibrinogen-like 2 (204834_at), score: 0.7 FHOD3formin homology 2 domain containing 3 (218980_at), score: -0.77 GPR116G protein-coupled receptor 116 (212950_at), score: 0.77 HS3ST3A1heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 (219985_at), score: -0.82 IFI27interferon, alpha-inducible protein 27 (202411_at), score: 0.78 JAG1jagged 1 (Alagille syndrome) (216268_s_at), score: 0.8 KRT18keratin 18 (201596_x_at), score: 0.74 LGALS3BPlectin, galactoside-binding, soluble, 3 binding protein (200923_at), score: 0.71 LIPGlipase, endothelial (219181_at), score: 0.85 LMO2LIM domain only 2 (rhombotin-like 1) (204249_s_at), score: 0.79 LPHN2latrophilin 2 (206953_s_at), score: 0.89 LRRC15leucine rich repeat containing 15 (213909_at), score: -0.8 LRRC17leucine rich repeat containing 17 (205381_at), score: 0.71 MBPmyelin basic protein (210136_at), score: 0.74 MEOX2mesenchyme homeobox 2 (206201_s_at), score: 1 MESTmesoderm specific transcript homolog (mouse) (202016_at), score: 0.8 MGPmatrix Gla protein (202291_s_at), score: 0.88 NDUFA4L2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 (218484_at), score: 0.93 NET1neuroepithelial cell transforming 1 (201830_s_at), score: -0.75 NKX3-1NK3 homeobox 1 (209706_at), score: -0.78 PDE10Aphosphodiesterase 10A (205501_at), score: 0.78 PLA2G16phospholipase A2, group XVI (209581_at), score: 0.78 PNMA2paraneoplastic antigen MA2 (209598_at), score: 0.74 PTPRZ1protein tyrosine phosphatase, receptor-type, Z polypeptide 1 (204469_at), score: 0.72 RELNreelin (205923_at), score: 0.78 RGS7regulator of G-protein signaling 7 (206290_s_at), score: 0.7 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: 0.71 SLC7A8solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 (216092_s_at), score: -0.82 SRGNserglycin (201859_at), score: 0.74 TINAGL1tubulointerstitial nephritis antigen-like 1 (219058_x_at), score: 0.76 TLR4toll-like receptor 4 (221060_s_at), score: 0.75 TMEM158transmembrane protein 158 (213338_at), score: -0.84 TNXAtenascin XA pseudogene (213451_x_at), score: 0.77 TNXBtenascin XB (216333_x_at), score: 0.77 ZNF423zinc finger protein 423 (214761_at), score: 0.88
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690223.cel | 3 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690256.cel | 6 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690392.cel | 14 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690472.cel | 17 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690480.cel | 18 | 5 | HGPS | hgu133a | HGPS | AG11498 |