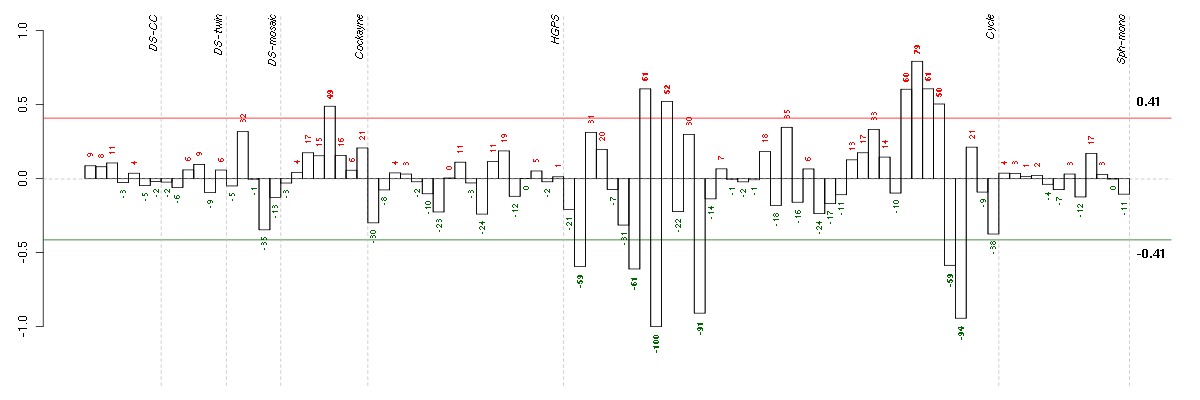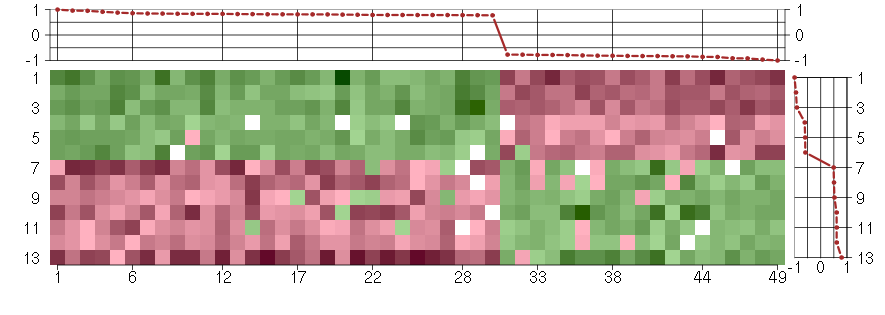

Under-expression is coded with green,
over-expression with red color.

biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
regulation of cell differentiation
Any process that modulates the frequency, rate or extent of cell differentiation, the process whereby relatively unspecialized cells acquire specialized structural and functional features.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of developmental process
Any process that modulates the frequency, rate or extent of development, the biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
regulation of developmental process
Any process that modulates the frequency, rate or extent of development, the biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cell differentiation
Any process that modulates the frequency, rate or extent of cell differentiation, the process whereby relatively unspecialized cells acquire specialized structural and functional features.
regulation of cell differentiation
Any process that modulates the frequency, rate or extent of cell differentiation, the process whereby relatively unspecialized cells acquire specialized structural and functional features.


ADORA2Badenosine A2b receptor (205891_at), score: 0.96 AHI1Abelson helper integration site 1 (221569_at), score: -0.86 AIM1absent in melanoma 1 (212543_at), score: 0.83 ANXA10annexin A10 (210143_at), score: -0.84 APOL6apolipoprotein L, 6 (219716_at), score: 0.78 BMP6bone morphogenetic protein 6 (206176_at), score: -0.86 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: 0.79 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.78 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.78 DAPK1death-associated protein kinase 1 (203139_at), score: 0.81 DCLK1doublecortin-like kinase 1 (205399_at), score: 0.81 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.81 DEPDC6DEP domain containing 6 (218858_at), score: 0.95 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.78 DOCK10dedicator of cytokinesis 10 (219279_at), score: -0.83 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.82 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: -0.77 FGF9fibroblast growth factor 9 (glia-activating factor) (206404_at), score: 0.82 GKglycerol kinase (207387_s_at), score: -0.96 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -1 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 1 IFIH1interferon induced with helicase C domain 1 (219209_at), score: 0.83 IRF1interferon regulatory factor 1 (202531_at), score: 0.81 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.83 KIAA1305KIAA1305 (220911_s_at), score: 0.78 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.83 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.81 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: -0.83 NCALDneurocalcin delta (211685_s_at), score: -0.78 NEFMneurofilament, medium polypeptide (205113_at), score: -0.91 NPTX1neuronal pentraxin I (204684_at), score: -0.8 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.83 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.77 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: 0.79 PCDH9protocadherin 9 (219737_s_at), score: -0.92 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.82 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: 0.77 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: 0.78 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: 0.91 SIX2SIX homeobox 2 (206510_at), score: 0.79 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.81 SMAD3SMAD family member 3 (218284_at), score: 0.84 SNNstannin (218032_at), score: 0.8 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.84 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.88 TLR3toll-like receptor 3 (206271_at), score: 0.85 TMEM2transmembrane protein 2 (218113_at), score: -0.79 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.81 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.78
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |