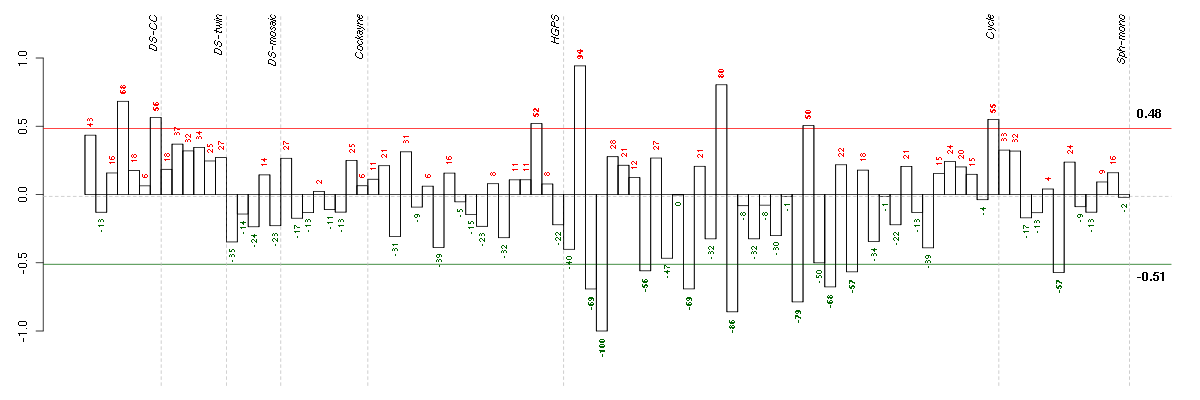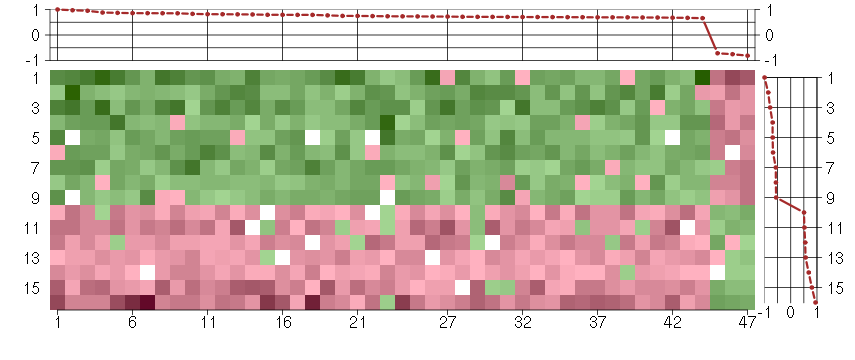

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
proteinaceous extracellular matrix
A layer consisting mainly of proteins (especially collagen) and glycosaminoglycans (mostly as proteoglycans) that forms a sheet underlying or overlying cells such as endothelial and epithelial cells. The proteins are secreted by cells in the vicinity.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: 0.86 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: 0.73 ADORA1adenosine A1 receptor (216220_s_at), score: 0.83 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: 0.69 ALDH3A1aldehyde dehydrogenase 3 family, memberA1 (205623_at), score: 0.71 ALPPL2alkaline phosphatase, placental-like 2 (216377_x_at), score: 0.68 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: 0.79 ATHL1ATH1, acid trehalase-like 1 (yeast) (219359_at), score: 0.71 C7orf58chromosome 7 open reading frame 58 (220032_at), score: -0.81 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 1 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.85 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: 0.73 CYTH4cytohesin 4 (219183_s_at), score: 0.67 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.8 EPM2Aepilepsy, progressive myoclonus type 2A, Lafora disease (laforin) (205231_s_at), score: 0.7 FBP1fructose-1,6-bisphosphatase 1 (209696_at), score: 0.66 FGL1fibrinogen-like 1 (205305_at), score: 0.75 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.79 FLRT3fibronectin leucine rich transmembrane protein 3 (219250_s_at), score: -0.75 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: 0.87 GPR144G protein-coupled receptor 144 (216289_at), score: 0.97 HAB1B1 for mucin (215778_x_at), score: 0.95 HAMPhepcidin antimicrobial peptide (220491_at), score: 0.71 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: 0.86 INE1inactivation escape 1 (non-protein coding) (207252_at), score: 0.73 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.85 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: 0.77 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.82 LOC80054hypothetical LOC80054 (220465_at), score: 0.69 MEOX1mesenchyme homeobox 1 (205619_s_at), score: 0.75 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.79 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: 0.74 NPEPL1aminopeptidase-like 1 (89476_r_at), score: 0.68 NXPH3neurexophilin 3 (221991_at), score: 0.7 OPRL1opiate receptor-like 1 (206564_at), score: 0.69 PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.81 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.88 PXNpaxillin (211823_s_at), score: -0.72 SAA4serum amyloid A4, constitutive (207096_at), score: 0.79 SEPT5septin 5 (209767_s_at), score: 0.7 SH2D3ASH2 domain containing 3A (222169_x_at), score: 0.7 SPATA1spermatogenesis associated 1 (221057_at), score: 0.72 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.71 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.81 TNK1tyrosine kinase, non-receptor, 1 (217149_x_at), score: 0.7 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: 0.72 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: 0.69
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |