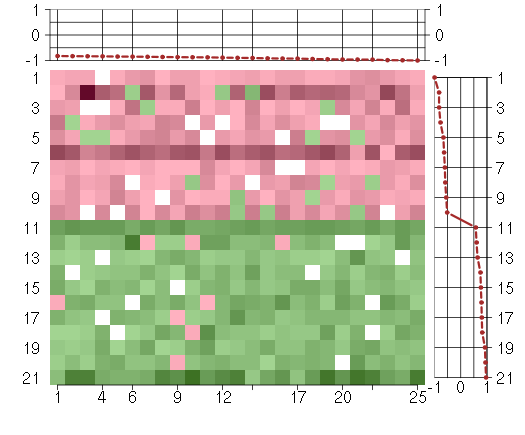

Under-expression is coded with green,
over-expression with red color.



ADORA1adenosine A1 receptor (216220_s_at), score: -0.99 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: -0.99 CATSPER2cation channel, sperm associated 2 (217588_at), score: -0.96 CXorf21chromosome X open reading frame 21 (220252_x_at), score: -0.92 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.96 DLX4distal-less homeobox 4 (208216_at), score: -0.87 F11coagulation factor XI (206610_s_at), score: -0.88 FAM75A3family with sequence similarity 75, member A3 (215935_at), score: -0.92 FOLH1folate hydrolase (prostate-specific membrane antigen) 1 (217487_x_at), score: -0.94 FUT2fucosyltransferase 2 (secretor status included) (210608_s_at), score: -0.85 GZMMgranzyme M (lymphocyte met-ase 1) (207460_at), score: -0.86 HAB1B1 for mucin (215778_x_at), score: -0.9 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: -0.95 LILRA5leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 (215838_at), score: -0.82 LRRN2leucine rich repeat neuronal 2 (216164_at), score: -0.85 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: -0.87 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: -0.93 NCRNA00092non-protein coding RNA 92 (215861_at), score: -0.9 PRINSpsoriasis associated RNA induced by stress (non-protein coding) (216051_x_at), score: -0.83 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (211349_at), score: -0.83 SLC26A10solute carrier family 26, member 10 (214951_at), score: -0.89 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: -0.86 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -1 TRA@T cell receptor alpha locus (216540_at), score: -0.96
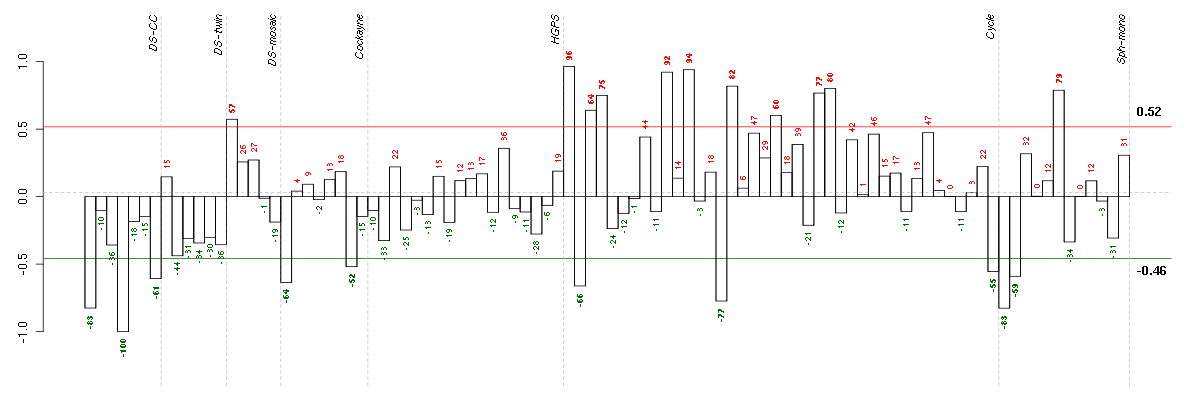
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486111.cel | 24 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |