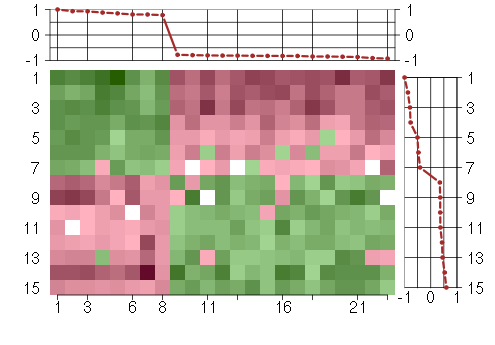

Under-expression is coded with green,
over-expression with red color.



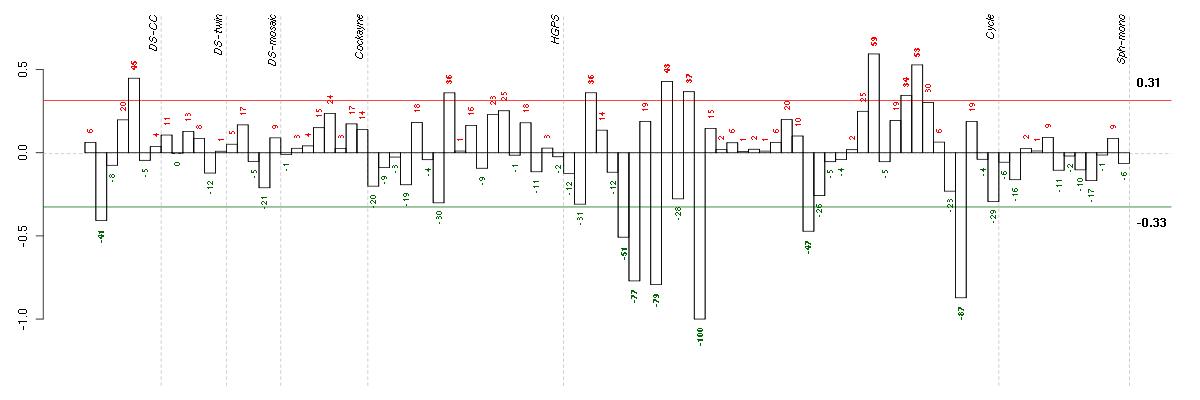
ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.85 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.82 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.81 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.88 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.81 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.78 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.79 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.82 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.8 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.82 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.84 KIAA1305KIAA1305 (220911_s_at), score: 0.8 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.82 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.93 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.93 LOC399491LOC399491 protein (214035_x_at), score: 0.81 NEFMneurofilament, medium polypeptide (205113_at), score: -0.91 NPIPnuclear pore complex interacting protein (204538_x_at), score: 1 PCDH9protocadherin 9 (219737_s_at), score: -0.92 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.78 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.84 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.86 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.85
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |