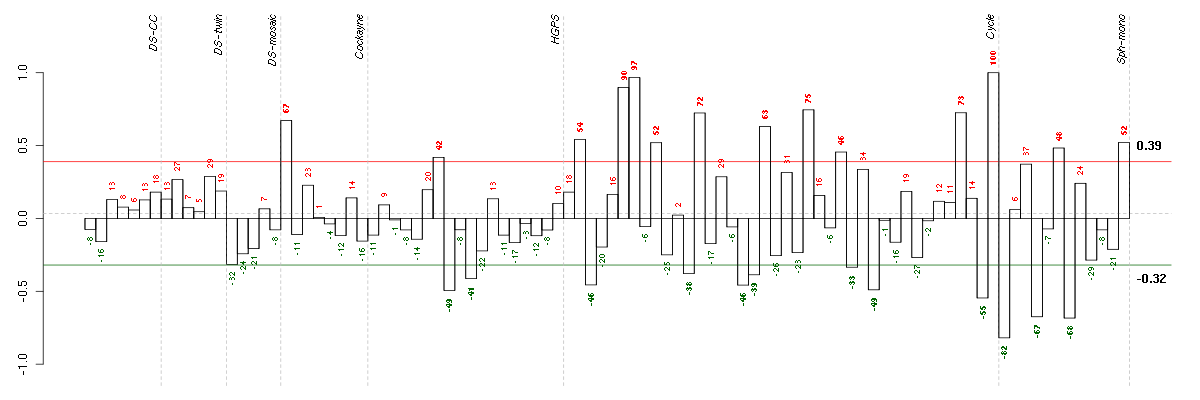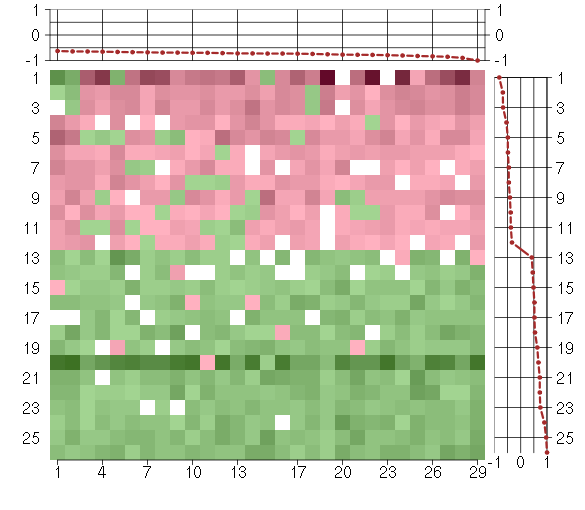

Under-expression is coded with green,
over-expression with red color.



B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (221484_at), score: -0.7 BIN1bridging integrator 1 (210201_x_at), score: -0.71 C20orf20chromosome 20 open reading frame 20 (218586_at), score: -0.68 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: -0.73 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.82 DOK4docking protein 4 (209691_s_at), score: -0.7 FAM134Cfamily with sequence similarity 134, member C (212697_at), score: -0.67 FBXL11F-box and leucine-rich repeat protein 11 (208989_s_at), score: -0.78 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: -0.69 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.73 FOXK2forkhead box K2 (203064_s_at), score: -1 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: -0.72 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.63 MYST2MYST histone acetyltransferase 2 (200049_at), score: -0.65 PCGF2polycomb group ring finger 2 (203793_x_at), score: -0.9 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: -0.72 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: -0.84 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: -0.78 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.64 SF1splicing factor 1 (208313_s_at), score: -0.74 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: -0.85 SPATA2spermatogenesis associated 2 (204433_s_at), score: -0.8 TAPBPTAP binding protein (tapasin) (208829_at), score: -0.79 TERF2telomeric repeat binding factor 2 (203611_at), score: -0.73 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: -0.69 TSKUtsukushin (218245_at), score: -0.77 UBAP1ubiquitin associated protein 1 (221490_at), score: -0.65 VAMP2vesicle-associated membrane protein 2 (synaptobrevin 2) (201557_at), score: -0.76 WDR42AWD repeat domain 42A (202249_s_at), score: -0.67
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |