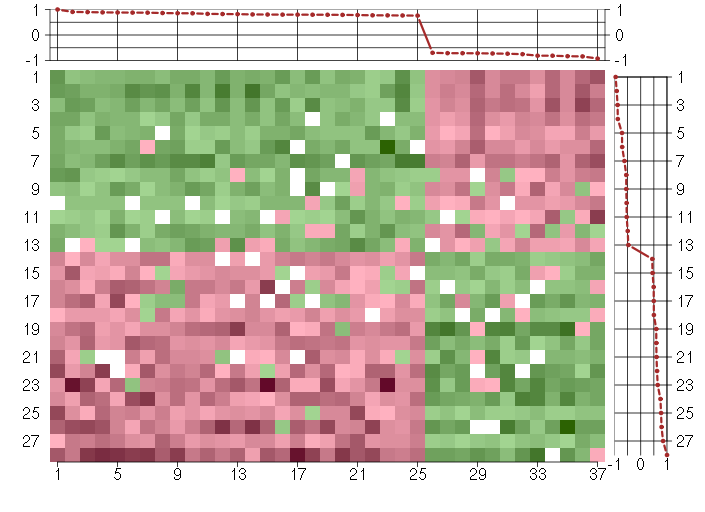

Under-expression is coded with green,
over-expression with red color.

defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
This term is the most general term possible


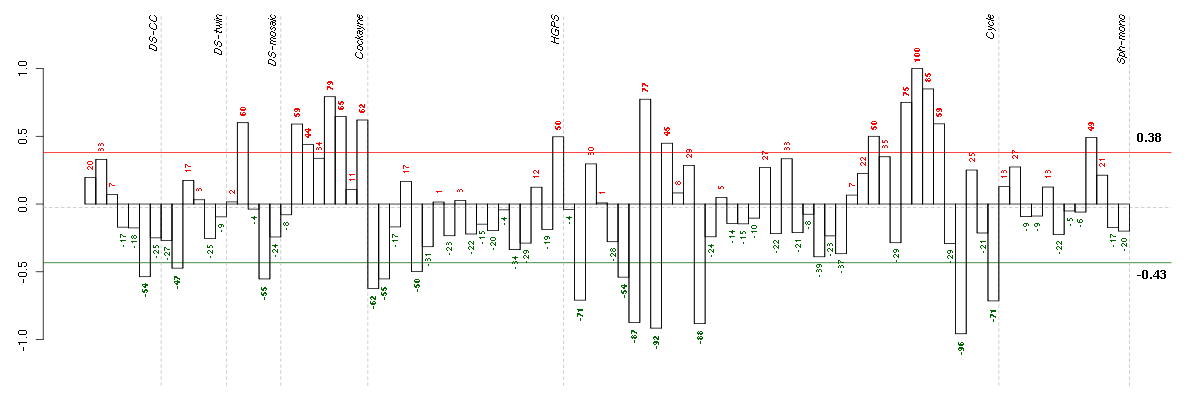
ACTC1actin, alpha, cardiac muscle 1 (205132_at), score: -0.81 ADORA2Badenosine A2b receptor (205891_at), score: 0.82 AIM1absent in melanoma 1 (212543_at), score: 0.77 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: 0.8 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: 0.88 ANGPTL2angiopoietin-like 2 (213004_at), score: 0.85 APOBEC3Gapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (204205_at), score: 0.79 APOL6apolipoprotein L, 6 (219716_at), score: 0.8 C5orf23chromosome 5 open reading frame 23 (219054_at), score: -0.71 DDX60DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 (218986_s_at), score: 0.76 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: -0.7 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.88 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: 0.77 IFIH1interferon induced with helicase C domain 1 (219209_at), score: 0.88 IRF1interferon regulatory factor 1 (202531_at), score: 0.86 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.79 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.81 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.75 MX2myxovirus (influenza virus) resistance 2 (mouse) (204994_at), score: 0.8 NEFMneurofilament, medium polypeptide (205113_at), score: -0.72 NMIN-myc (and STAT) interactor (203964_at), score: 0.85 NTMneurotrimin (222020_s_at), score: -0.73 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.89 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.92 PARP12poly (ADP-ribose) polymerase family, member 12 (218543_s_at), score: 0.77 PCDH9protocadherin 9 (219737_s_at), score: -0.84 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: 0.82 SECTM1secreted and transmembrane 1 (213716_s_at), score: 0.78 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: -0.73 SMAD3SMAD family member 3 (218284_at), score: 0.81 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.9 TLR3toll-like receptor 3 (206271_at), score: 1 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.8 TSPAN13tetraspanin 13 (217979_at), score: -0.83 UBE2L6ubiquitin-conjugating enzyme E2L 6 (201649_at), score: 0.83 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.91 XYLT1xylosyltransferase I (213725_x_at), score: -0.71
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| E-GEOD-3860-raw-cel-1561690215.cel | 2 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690480.cel | 18 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |