Previous module |
Next module
Module #545, TG: 3, TC: 2, 73 probes, 72 Entrez genes, 6 conditions


HELP
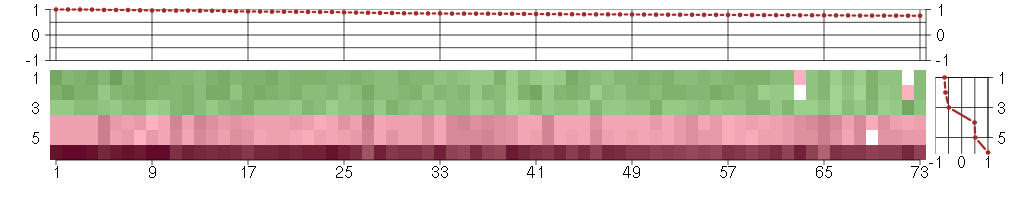
The image plot shows the color-coded level of gene expression, for the
genes and conditions in a given transcription module. The genes are on
the horizontal, the conditions on the vertical axis.
The genes are ordered according to their ISA gene scores, similarly
the conditions are ordered according to their condition scores. The
score of a gene means the «degree of inclusion» in
the module: a high score gene is essential in the module.
Condition scores can also be negative, that means that the genes of
the module are all down-regulated in the condition. Here the absolute
value of the score gives the «degree of inclusion».
The plots above and beside the expression matrix show the gene scores
and condition scores, respectively.
Note that the plot is interactive, you can see the name of the gene
and condition under the mouse cursor.
The expression matrix was normalized to have mean zero and standard
deviation one for every gene separately across all conditions
(i.e. not just for the conditions in the module).
— Click on the Help button again to close this help window.

Under-expression is coded with green,
over-expression with red color.
Help |
Hide |
Top
Help |
Show |
Top
The GO tree — Biological processes
HELP
This is one of three sections showing Gene Ontology enrichment of the
current module: in this case for biological processes.
The graph shows the hierarchy of the GO categories, their enrichment
for the current module is color coded, and the blue number beside the
category is the minus log ten p-value of the enrichment. (Calculated
using the standard hypergeometric test.) The color of the arrows code
«is a» (cyan) and «part of» relationships.
The tree was built the following way. First all GO terms with more
significant enrichment p-value than 0.05 were collected. Then all
paths from these terms to the root node of the GO tree were included
too. If a GO term is included more than once in the tree, then the
green numbers show 1) the id of the node, this makes it easier to find
other appereances of the term, and 2) the number of appearences.
Note that the same GO category might show up on the graph many
times. This is because the GO was «straightened» for this
graph, i.e. if there are more paths from a GO term to the root node of
the tree, all of them are included. The green numbers
Move the mouse cursor over the terms to get their definition. Clicking
on them takes you to the corresponding Gene Ontology web page.
If you cannot see a graph here at all, that means that there were no
significantly enriched GO categories, at the 0.05 level.
— Click on the Help button again to close this help window.

Help |
Hide |
Top
Help |
Show |
Top
The GO tree — Cellular Components
HELP
This is one of three sections showing Gene Ontology enrichment of the
current module: in this case for cellular components.
The graph shows the hierarchy of the GO categories, their enrichment
for the current module is color coded, and the blue number beside the
category is the minus log ten p-value of the enrichment. (Calculated
using the standard hypergeometric test.) The color of the arrows code
«is a» (cyan) and «part of» relationships.
The tree was built the following way. First all GO terms with more
significant enrichment p-value than 0.05 were collected. Then all
paths from these terms to the root node of the GO tree were included
too. If a GO term is included more than once in the tree, then the
green numbers show 1) the id of the node, this makes it easier to find
other appereances of the term, and 2) the number of appearences.
Note that the same GO category might show up on the graph many
times. This is because the GO was «straightened» for this
graph, i.e. if there are more paths from a GO term to the root node of
the tree, all of them are included. The green numbers
Move the mouse cursor over the terms to get their definition. Clicking
on them takes you to the corresponding Gene Ontology web page.
If you cannot see a graph here at all, that means that there were no
significantly enriched GO categories, at the 0.05 level.
— Click on the Help button again to close this help window.

Help |
Hide |
Top
Help |
Show |
Top
The GO tree — Molecular Function
HELP
This is one of three sections showing Gene Ontology enrichment of the
current module: in this case for molecular function.
The graph shows the hierarchy of the GO categories, their enrichment
for the current module is color coded, and the blue number beside the
category is the minus log ten p-value of the enrichment. (Calculated
using the standard hypergeometric test.) The color of the arrows code
«is a» (cyan) and «part of» relationships.
The tree was built the following way. First all GO terms with more
significant enrichment p-value than 0.05 were collected. Then all
paths from these terms to the root node of the GO tree were included
too. If a GO term is included more than once in the tree, then the
green numbers show 1) the id of the node, this makes it easier to find
other appereances of the term, and 2) the number of appearences.
Note that the same GO category might show up on the graph many
times. This is because the GO was «straightened» for this
graph, i.e. if there are more paths from a GO term to the root node of
the tree, all of them are included. The green numbers
Move the mouse cursor over the terms to get their definition. Clicking
on them takes you to the corresponding Gene Ontology web page.
If you cannot see a graph here at all, that means that there were no
significantly enriched GO categories, at the 0.05 level.
— Click on the Help button again to close this help window.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
metal ion binding
Interacting selectively with any metal ion.
ion binding
Interacting selectively with ions, charged atoms or groups of atoms.
cation binding
Interacting selectively with cations, charged atoms or groups of atoms with a net positive charge.
transition metal ion binding
Interacting selectively with a transition metal ions; a transition metal is an element whose atom has an incomplete d-subshell of extranuclear electrons, or which gives rise to a cation or cations with an incomplete d-subshell. Transition metals often have more than one valency state. Biologically relevant transition metals include vanadium, manganese, iron, copper, cobalt, nickel, molybdenum and silver.
all
This term is the most general term possible
transition metal ion binding
Interacting selectively with a transition metal ions; a transition metal is an element whose atom has an incomplete d-subshell of extranuclear electrons, or which gives rise to a cation or cations with an incomplete d-subshell. Transition metals often have more than one valency state. Biologically relevant transition metals include vanadium, manganese, iron, copper, cobalt, nickel, molybdenum and silver.
Help |
Hide |
Top
Help |
Show |
Top
GO BP test for over-representation
HELP
List of all enriched GO categories (biological processes), at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module annotated with the given GO term, just by chance.
- Count
is the number of genes in the module annotated with the given GO
term.
- Size is the total number of genes (in our universe)
annotated with the GO term.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
GO category.
Clicking on the GO identifiers takes you to the Gene Ontology web
pages.
— Click on the Help button again to close this help window.
Help |
Hide |
Top
Help |
Show |
Top
GO CC test for over-representation
HELP
List of all enriched GO categories (cellular components), at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module annotated with the given GO term, just by chance.
- Count
is the number of genes in the module annotated with the given GO
term.
- Size is the total number of genes (in our universe)
annotated with the GO term.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
GO category.
Clicking on the GO identifiers takes you to the Gene Ontology web
pages.
— Click on the Help button again to close this help window.
Help |
Hide |
Top
Help |
Show |
Top
GO MF test for over-representation
HELP
List of all enriched GO categories (molecular function), at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module annotated with the given GO term, just by chance.
- Count
is the number of genes in the module annotated with the given GO
term.
- Size is the total number of genes (in our universe)
annotated with the GO term.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
GO category.
Clicking on the GO identifiers takes you to the Gene Ontology web
pages.
— Click on the Help button again to close this help window.
| Id | Pvalue | ExpCount | Count | Size | Term |
| GO:0046914 | 6.002e-03 | 9.389 | 22
CBFA2T2, CISD3, CYP2B6, FBXO41, LIMK1, MEFV, METAP2, PCGF2, POGZ, POLR1B, SCD5, SLC30A5, TIMM8A, ZBTB39, ZC3H7B, ZNF492, ZNF528, ZNF552, ZNF611, ZNF639, ZNF816A, ZSCAN12 | 1404 | transition metal ion binding |
| GO:0008270 | 1.114e-02 | 7.691 | 19
CBFA2T2, CISD3, FBXO41, LIMK1, MEFV, PCGF2, POGZ, POLR1B, SLC30A5, TIMM8A, ZBTB39, ZC3H7B, ZNF492, ZNF528, ZNF552, ZNF611, ZNF639, ZNF816A, ZSCAN12 | 1150 | zinc ion binding |
| GO:0003677 | 3.837e-02 | 7.992 | 18
CBFA2T2, G3BP1, HEY1, JARID2, PCGF2, PMS2CL, POGZ, POLR1B, PPARGC1A, XRCC2, ZBTB39, ZNF492, ZNF528, ZNF552, ZNF611, ZNF639, ZNF816A, ZSCAN12 | 1195 | DNA binding |
Help |
Hide |
Top
Help |
Show |
Top
KEGG Pathway test for over-representation
HELP
List of all enriched KEGG pathways, at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module annotated with the given KEGG pathway, just by chance.
- Count
is the number of genes in the module annotated with the given KEGG
pathway.
- Size is the total number of genes (in our universe)
annotated with the KEGG pathway.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
KEGG pathway.
Clicking on the KEGG identifiers takes you to the KEGG web site.
— Click on the Help button again to close this help window.
Help |
Hide |
Top
Help |
Show |
Top
miRNA test for over-representation
HELP
List of all enriched miRNA families, at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module regulated by the given miRNA family, just by chance.
- Count
is the number of genes in the module regulated by the given miRNA
family.
- Size is the total number of genes (in our universe)
regulated with the given miRNA family.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
miRNA family.
The miRNA regulation data was taken from the TargetScan database.
(Only the conserved sites were used for the current analysis.)
Clicking on the miRNA names takes you to the TargetScan web site.
— Click on the Help button again to close this help window.
| Id | Pvalue | ExpCount | Count | Size |
| miR-138 | 4.029e-02 | 1.474 | 9
C11orf57, CBFA2T2, FBXO41, JARID2, LIMK1, MAP2K7, PIP4K2C, PPARGC1A, ZC3H7B | 253 |
Help |
Hide |
Top
Help |
Show |
Top
Chromosome test for over-representation
HELP
List of all enriched Chromosomes, at the 0.05
p-value level.
The columns:
- ExpCount is the expected number of genes in the
module on the given chromosome, just by chance.
- Count
is the number of genes in the module on the given chromosome.
- Size is the total number of genes (in our universe)
on the given chromosome.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
chromosome.
— Click on the Help button again to close this help window.
HELP
A list of all genes in the current module, in alphabetical order. The
size of the text corresponds to the gene scores.
Note that some gene symbols may show up more than once, if many
probes match the same Entrez gene.
Genes with no Entrez mapping are given separately, with their
Affymetrics probe ID.
— Click on the Help button again to close this help window.
Entrez genes
ALDOAP2aldolase A, fructose-bisphosphate pseudogene 2 (211617_at), score: 0.79
ALMS1Alstrom syndrome 1 (214707_x_at), score: 0.9
APBB2amyloid beta (A4) precursor protein-binding, family B, member 2 (212972_x_at), score: 0.96
C11orf57chromosome 11 open reading frame 57 (218314_s_at), score: 0.76
C17orf86chromosome 17 open reading frame 86 (221621_at), score: 0.98
CBFA2T2core-binding factor, runt domain, alpha subunit 2; translocated to, 2 (207625_s_at), score: 0.83
CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213183_s_at), score: 0.95
CISD3CDGSH iron sulfur domain 3 (213551_x_at), score: 0.82
CYP2B6cytochrome P450, family 2, subfamily B, polypeptide 6 (217133_x_at), score: 0.8
DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (219290_x_at), score: 0.91
DDX6DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 (204909_at), score: 0.76
DNAH3dynein, axonemal, heavy chain 3 (220725_x_at), score: 1
EPB41L1erythrocyte membrane protein band 4.1-like 1 (212339_at), score: 0.96
FAM134Afamily with sequence similarity 134, member A (222129_at), score: 0.84
FBXO41F-box protein 41 (44040_at), score: 0.84
FBXW12F-box and WD repeat domain containing 12 (215600_x_at), score: 0.83
FCARFc fragment of IgA, receptor for (211305_x_at), score: 0.86
FLJ11292hypothetical protein FLJ11292 (220828_s_at), score: 1
FLJ23172hypothetical LOC389177 (217016_x_at), score: 0.8
FNBP4formin binding protein 4 (212232_at), score: 0.77
G3BP1GTPase activating protein (SH3 domain) binding protein 1 (222187_x_at), score: 1
GCLCglutamate-cysteine ligase, catalytic subunit (202922_at), score: 0.75
HCG2P7HLA complex group 2 pseudogene 7 (216229_x_at), score: 0.91
HEY1hairy/enhancer-of-split related with YRPW motif 1 (44783_s_at), score: 0.93
IBD12Inflammatory bowel disease 12 (215373_x_at), score: 0.87
JARID2jumonji, AT rich interactive domain 2 (203297_s_at), score: 0.79
LIMK1LIM domain kinase 1 (204357_s_at), score: 0.77
LOC100134401hypothetical protein LOC100134401 (213605_s_at), score: 0.8
LOC643313similar to hypothetical protein LOC284701 (211050_x_at), score: 0.94
LOC647070hypothetical LOC647070 (215467_x_at), score: 0.92
MAP2K7mitogen-activated protein kinase kinase 7 (216206_x_at), score: 0.85
MEFVMediterranean fever (208262_x_at), score: 0.78
METAP2methionyl aminopeptidase 2 (202015_x_at), score: 0.85
METTL7Amethyltransferase like 7A (211424_x_at), score: 0.9
MFSD11major facilitator superfamily domain containing 11 (221192_x_at), score: 0.79
MRPL44mitochondrial ribosomal protein L44 (218202_x_at), score: 0.78
PCGF2polycomb group ring finger 2 (203793_x_at), score: 0.84
PIP4K2Cphosphatidylinositol-5-phosphate 4-kinase, type II, gamma (218942_at), score: 0.76
PMS2CLPMS2 C-terminal like pseudogene (221206_at), score: 0.77
POGZpogo transposable element with ZNF domain (215281_x_at), score: 0.81
POLR1Bpolymerase (RNA) I polypeptide B, 128kDa (220113_x_at), score: 0.85
POM121L9PPOM121 membrane glycoprotein-like 9 (rat) pseudogene (222253_s_at), score: 0.76
PPARGC1Aperoxisome proliferator-activated receptor gamma, coactivator 1 alpha (219195_at), score: 0.87
PRINSpsoriasis associated RNA induced by stress (non-protein coding) (216051_x_at), score: 0.82
RPL21P37ribosomal protein L21 pseudogene 37 (216479_at), score: 0.89
RPL35Aribosomal protein L35a (215208_x_at), score: 0.81
SAPS2SAPS domain family, member 2 (202792_s_at), score: 0.84
SCD5stearoyl-CoA desaturase 5 (220232_at), score: 0.81
SH3BP2SH3-domain binding protein 2 (217257_at), score: 0.83
SH3GL3SH3-domain GRB2-like 3 (211565_at), score: 0.97
SKP1S-phase kinase-associated protein 1 (200719_at), score: 0.93
SLC30A5solute carrier family 30 (zinc transporter), member 5 (220181_x_at), score: 0.98
SLC35F2solute carrier family 35, member F2 (218826_at), score: 0.78
SPATA2spermatogenesis associated 2 (204433_s_at), score: 0.85
SPG21spastic paraplegia 21 (autosomal recessive, Mast syndrome) (215383_x_at), score: 0.79
SPINLW1serine peptidase inhibitor-like, with Kunitz and WAP domains 1 (eppin) (206318_at), score: 0.96
SPNsialophorin (206057_x_at), score: 0.96
SPTLC3serine palmitoyltransferase, long chain base subunit 3 (220456_at), score: 0.98
TCTN2tectonic family member 2 (206438_x_at), score: 0.81
TIMM8Atranslocase of inner mitochondrial membrane 8 homolog A (yeast) (210800_at), score: 1
UBQLN4ubiquilin 4 (222252_x_at), score: 0.88
VAMP2vesicle-associated membrane protein 2 (synaptobrevin 2) (201557_at), score: 0.95
XRCC2X-ray repair complementing defective repair in Chinese hamster cells 2 (207598_x_at), score: 0.8
ZBTB39zinc finger and BTB domain containing 39 (205256_at), score: 0.78
ZC3H7Bzinc finger CCCH-type containing 7B (206169_x_at), score: 0.92
ZNF492zinc finger protein 492 (215532_x_at), score: 0.76
ZNF528zinc finger protein 528 (215019_x_at), score: 0.81
ZNF552zinc finger protein 552 (219741_x_at), score: 0.78
ZNF611zinc finger protein 611 (208137_x_at), score: 0.79
ZNF639zinc finger protein 639 (218413_s_at), score: 0.78
ZNF816Azinc finger protein 816A (217541_x_at), score: 0.91
ZSCAN12zinc finger and SCAN domain containing 12 (206507_at), score: 0.81
Non-Entrez genes
205256_atUnknown, score: 0.82
HELP
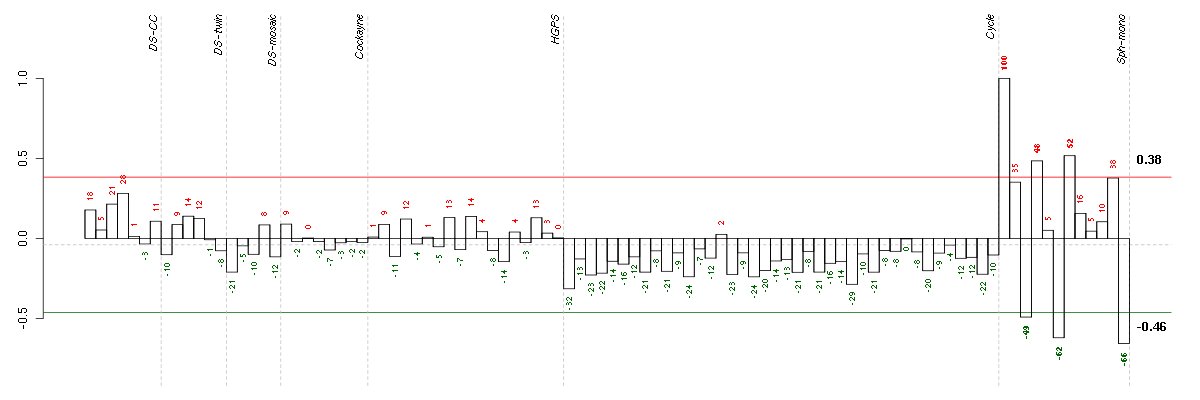
Conditions in the module, given in the same order as on the expression
plot above. Red color means over-expression, green under-expression in
the given condition.
The barplot below shows the condition (sample) scores. A separate bar
is shown for each sample, its height is the corresponding score of the
sample in the module. The red and green numbers on the bars are the
sample scores expressed in percents, i.e. 100% is 1.0.
The red and green lines show the module thresholds, samples above
the red line and below the green line are included in the module.
The different experiments that were part of the study, are separated
by dashed vertical lines.
— Click on the Help button again to close this help window.
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |



© 2008-2010 Computational Biology Group, Department of Medical Genetics,
University of Lausanne, Switzerland