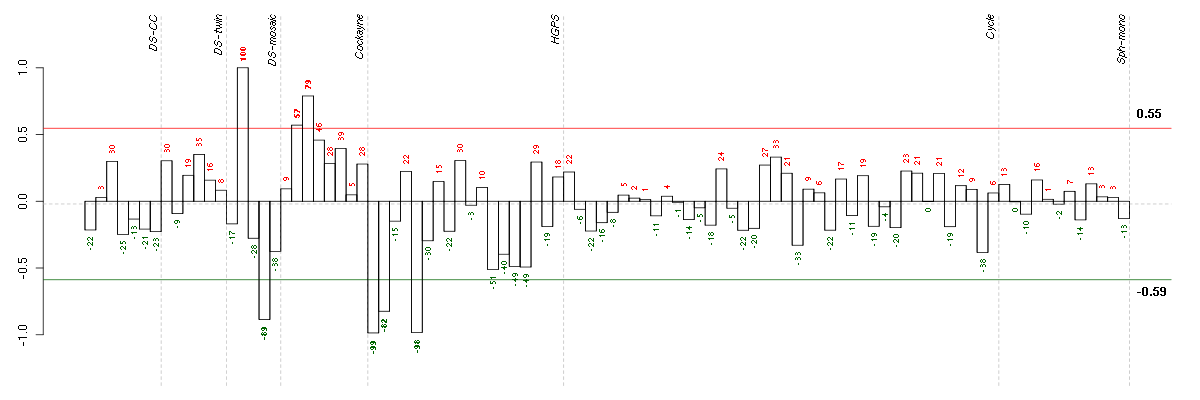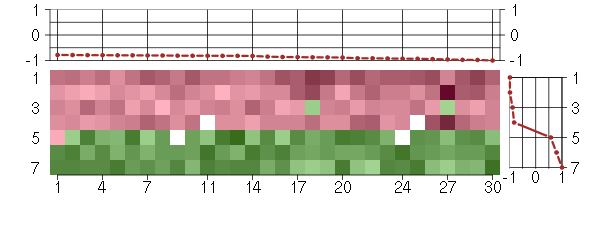

Under-expression is coded with green,
over-expression with red color.

cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
biological adhesion
The attachment of a cell or organism to a substrate or other organism.
all
This term is the most general term possible
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.


ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: -1 ARPC4actin related protein 2/3 complex, subunit 4, 20kDa (211672_s_at), score: -0.93 BCL2L1BCL2-like 1 (215037_s_at), score: -0.81 C19orf6chromosome 19 open reading frame 6 (213986_s_at), score: -0.82 C5orf23chromosome 5 open reading frame 23 (219054_at), score: -0.97 CGBchorionic gonadotropin, beta polypeptide (205387_s_at), score: -0.88 COMPcartilage oligomeric matrix protein (205713_s_at), score: -0.98 CRLF1cytokine receptor-like factor 1 (206315_at), score: -0.88 EZRezrin (208621_s_at), score: -0.86 FSCN1fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) (210933_s_at), score: -0.82 GRAMD3GRAM domain containing 3 (218706_s_at), score: -0.91 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: -0.87 KRTAP1-1keratin associated protein 1-1 (220976_s_at), score: -0.96 LOXL2lysyl oxidase-like 2 (202997_s_at), score: -0.92 MMP14matrix metallopeptidase 14 (membrane-inserted) (202827_s_at), score: -0.81 MYO10myosin X (201976_s_at), score: -0.8 NTMneurotrimin (222020_s_at), score: -0.88 PFKLphosphofructokinase, liver (201102_s_at), score: -0.79 PLEC1plectin 1, intermediate filament binding protein 500kDa (216971_s_at), score: -0.93 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: -0.8 PTMSparathymosin (218045_x_at), score: -0.78 PXNpaxillin (211823_s_at), score: -0.8 SEC14L2SEC14-like 2 (S. cerevisiae) (204541_at), score: -0.8 SH3PXD2ASH3 and PX domains 2A (213252_at), score: -0.79 SLC5A3solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 (213164_at), score: -0.85 SRPRsignal recognition particle receptor (docking protein) (200917_s_at), score: -0.79 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (217790_s_at), score: -0.91 TNFRSF10Dtumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain (210654_at), score: -0.94 XYLT1xylosyltransferase I (213725_x_at), score: -0.82 ZNF185zinc finger protein 185 (LIM domain) (203585_at), score: -0.81
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| E-GEOD-3860-raw-cel-1561690215.cel | 2 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |