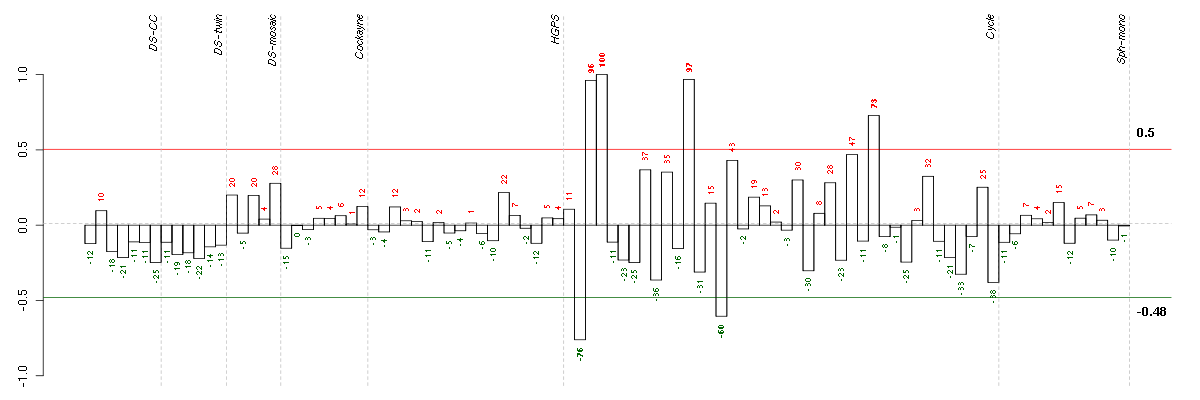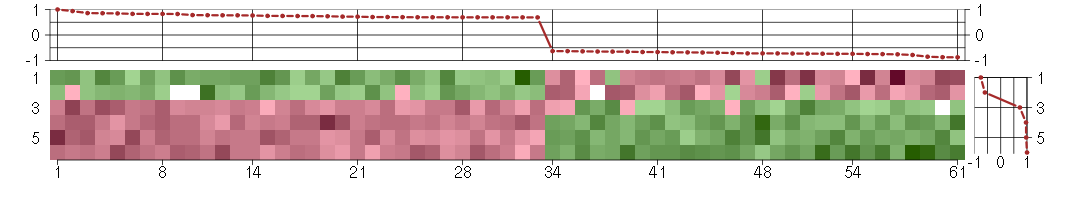

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: -0.76 ADORA1adenosine A1 receptor (216220_s_at), score: -0.68 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.72 C2orf37chromosome 2 open reading frame 37 (220172_at), score: 0.75 C7orf58chromosome 7 open reading frame 58 (220032_at), score: 0.7 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: -0.87 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.72 DENND3DENN/MADD domain containing 3 (212974_at), score: 0.74 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: -0.71 EMCNendomucin (219436_s_at), score: 1 FBP1fructose-1,6-bisphosphatase 1 (209696_at), score: -0.78 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: -0.72 FLRT3fibronectin leucine rich transmembrane protein 3 (219250_s_at), score: 0.77 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: -0.88 GDF9growth differentiation factor 9 (221314_at), score: -0.67 GPR56G protein-coupled receptor 56 (212070_at), score: 0.69 HAB1B1 for mucin (215778_x_at), score: -0.75 HIST1H4Ghistone cluster 1, H4g (208551_at), score: -0.73 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.77 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: 0.69 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: -0.75 IL9Rinterleukin 9 receptor (217212_s_at), score: -0.74 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: 0.69 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: -0.66 JAK2Janus kinase 2 (a protein tyrosine kinase) (205842_s_at), score: 0.69 JUPjunction plakoglobin (201015_s_at), score: 0.75 KCTD15potassium channel tetramerisation domain containing 15 (218553_s_at), score: -0.67 KCTD2potassium channel tetramerisation domain containing 2 (212564_at), score: 0.69 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: -0.68 LOC391132similar to hCG2041276 (216177_at), score: -0.65 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.7 MEOX1mesenchyme homeobox 1 (205619_s_at), score: -0.73 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: -0.69 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: 0.7 MTMR11myotubularin related protein 11 (205076_s_at), score: 0.82 NCOA2nuclear receptor coactivator 2 (205732_s_at), score: 0.77 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.85 NF1neurofibromin 1 (211094_s_at), score: 0.85 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.94 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.83 PCNXpecanex homolog (Drosophila) (213159_at), score: 0.69 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.78 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: -0.85 PXNpaxillin (211823_s_at), score: 0.83 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: 0.83 REG3Aregenerating islet-derived 3 alpha (205815_at), score: -0.65 RP4-691N24.1ninein-like (207705_s_at), score: 0.72 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: 0.86 SENP7SUMO1/sentrin specific peptidase 7 (220735_s_at), score: 0.74 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: 0.7 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: -0.63 STIM1stromal interaction molecule 1 (202764_at), score: 0.78 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.74 TGM4transglutaminase 4 (prostate) (217566_s_at), score: -0.73 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.7 TNK1tyrosine kinase, non-receptor, 1 (217149_x_at), score: -0.63 TOM1L2target of myb1-like 2 (chicken) (214840_at), score: 0.69 TULP2tubby like protein 2 (206733_at), score: -0.74 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.72 ZNF652zinc finger protein 652 (205594_at), score: -0.64
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |