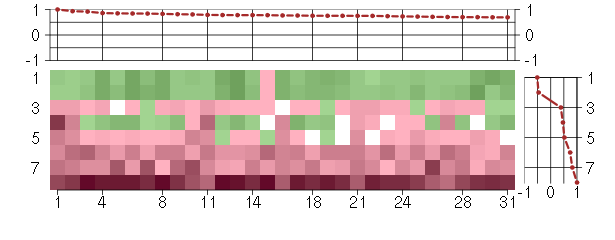

Under-expression is coded with green,
over-expression with red color.

defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
behavior
The specific actions or reactions of an organism in response to external or internal stimuli. Patterned activity of a whole organism in a manner dependent upon some combination of that organism's internal state and external conditions.
locomotory behavior
The specific movement from place to place of an organism in response to external or internal stimuli. Locomotion of a whole organism in a manner dependent upon some combination of that organism's internal state and external conditions.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
This term is the most general term possible
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04060 | 1.131e-02 | 0.5562 | 5 | 115 | Cytokine-cytokine receptor interaction |
ABCG1ATP-binding cassette, sub-family G (WHITE), member 1 (204567_s_at), score: 0.84 ADORA2Aadenosine A2a receptor (205013_s_at), score: 0.92 ANKHankylosis, progressive homolog (mouse) (220076_at), score: 0.7 ANKRD10ankyrin repeat domain 10 (218093_s_at), score: 0.69 APODapolipoprotein D (201525_at), score: 1 BHLHB9basic helix-loop-helix domain containing, class B, 9 (213709_at), score: 0.76 CHI3L1chitinase 3-like 1 (cartilage glycoprotein-39) (209395_at), score: 0.78 CHST2carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 (203921_at), score: 0.7 CXCL1chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) (204470_at), score: 0.75 CXCL2chemokine (C-X-C motif) ligand 2 (209774_x_at), score: 0.93 CXCL3chemokine (C-X-C motif) ligand 3 (207850_at), score: 0.79 DKFZp686O1327hypothetical gene supported by BC043549; BX648102 (216877_at), score: 0.75 GPM6Bglycoprotein M6B (209167_at), score: 0.81 HNRNPDheterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) (213359_at), score: 0.75 IL1RAPinterleukin 1 receptor accessory protein (205227_at), score: 0.72 IL1RNinterleukin 1 receptor antagonist (212657_s_at), score: 0.81 IL24interleukin 24 (206569_at), score: 0.71 INSRinsulin receptor (213792_s_at), score: 0.86 ITPKBinositol 1,4,5-trisphosphate 3-kinase B (203723_at), score: 0.69 NACAnascent polypeptide-associated complex alpha subunit (222018_at), score: 0.74 PELI2pellino homolog 2 (Drosophila) (219132_at), score: 0.84 RGS5regulator of G-protein signaling 5 (218353_at), score: 0.85 SH3BP2SH3-domain binding protein 2 (217257_at), score: 0.78 SOD2superoxide dismutase 2, mitochondrial (221477_s_at), score: 0.77 SYCP1synaptonemal complex protein 1 (206740_x_at), score: 0.75 TRIM36tripartite motif-containing 36 (219736_at), score: 0.78 TRPV1transient receptor potential cation channel, subfamily V, member 1 (219632_s_at), score: 0.75 UGT2B28UDP glucuronosyltransferase 2 family, polypeptide B28 (211682_x_at), score: 0.73 USP6ubiquitin specific peptidase 6 (Tre-2 oncogene) (206405_x_at), score: 0.7 ZBTB39zinc finger and BTB domain containing 39 (205256_at), score: 0.83 ZNF770zinc finger protein 770 (220608_s_at), score: 0.78
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
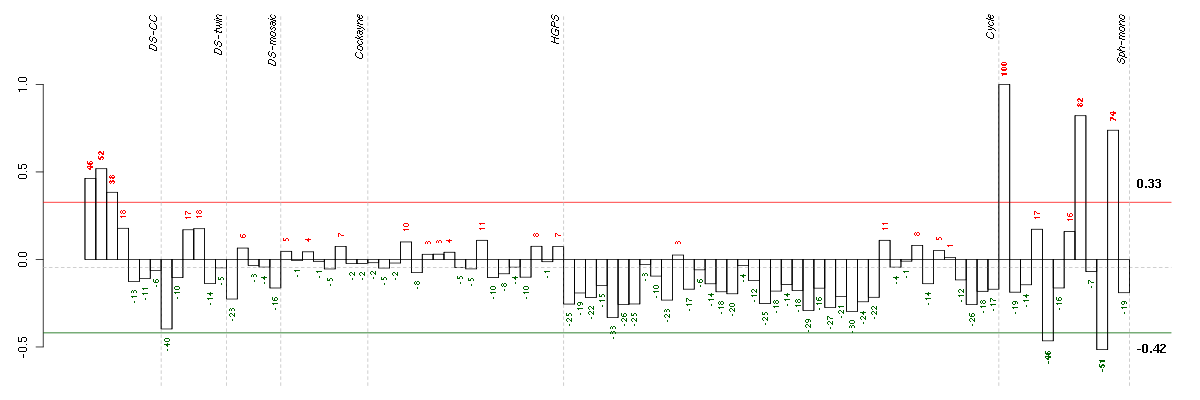
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |