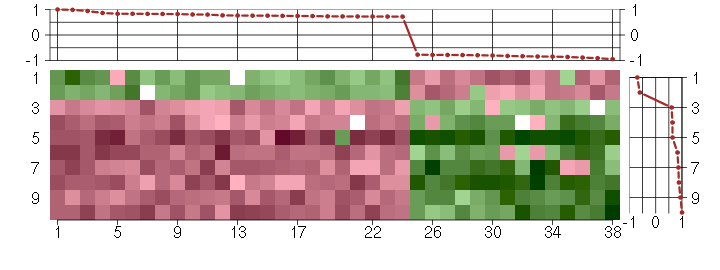

Under-expression is coded with green,
over-expression with red color.



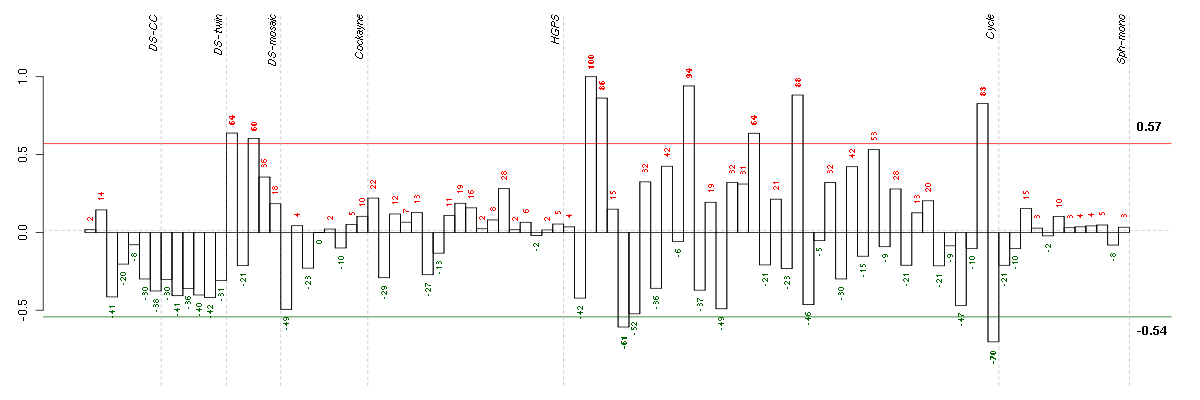
ATF5activating transcription factor 5 (204999_s_at), score: 0.73 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.74 C1orf69chromosome 1 open reading frame 69 (215490_at), score: -0.79 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: -0.82 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.95 DENND3DENN/MADD domain containing 3 (212974_at), score: 0.76 FBXO2F-box protein 2 (219305_x_at), score: -0.79 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.83 HAB1B1 for mucin (215778_x_at), score: -0.85 HIST1H4Dhistone cluster 1, H4d (208076_at), score: -0.83 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.76 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.8 INPP1inositol polyphosphate-1-phosphatase (202794_at), score: 0.73 JUPjunction plakoglobin (201015_s_at), score: 0.75 LEPREL2leprecan-like 2 (204854_at), score: 0.82 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.86 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.8 OR7E47Polfactory receptor, family 7, subfamily E, member 47 pseudogene (216698_x_at), score: -0.77 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.72 PHF7PHD finger protein 7 (215622_x_at), score: -0.78 PRKCBprotein kinase C, beta (209685_s_at), score: -0.89 PRKCDprotein kinase C, delta (202545_at), score: 0.77 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: -0.84 PXNpaxillin (211823_s_at), score: 0.94 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.72 RASA3RAS p21 protein activator 3 (206220_s_at), score: 0.82 RLN1relaxin 1 (211753_s_at), score: -0.86 SLC9A3R2solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 (209830_s_at), score: 0.75 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.76 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.83 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.72 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.83 TGIF2TGFB-induced factor homeobox 2 (216262_s_at), score: -0.8 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: -0.78 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.99 VEGFBvascular endothelial growth factor B (203683_s_at), score: 1 ZFHX3zinc finger homeobox 3 (208033_s_at), score: 0.73
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |