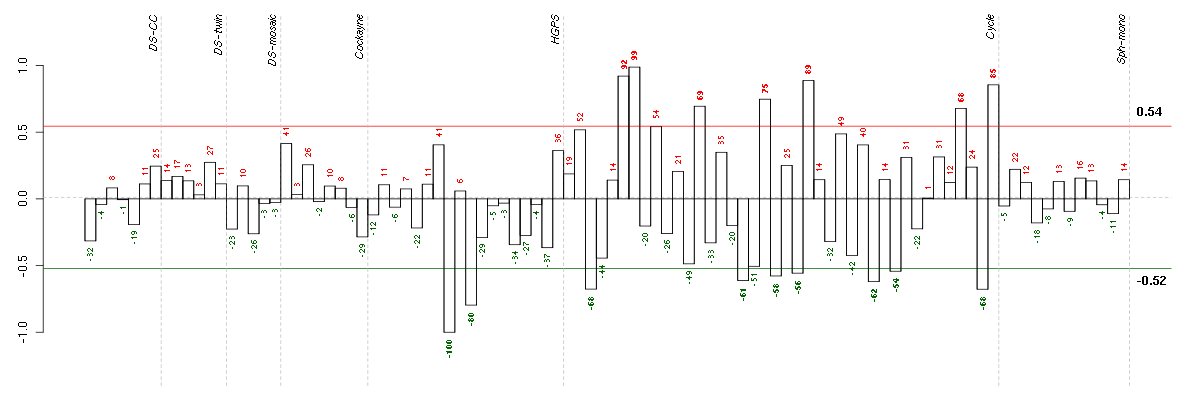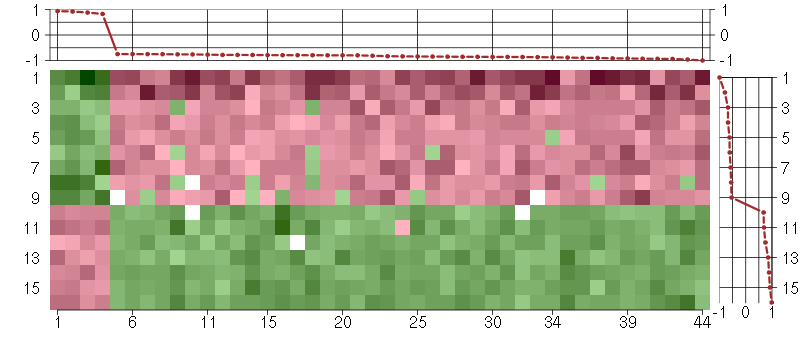

Under-expression is coded with green,
over-expression with red color.



ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.85 ATN1atrophin 1 (40489_at), score: -0.93 BAIAP2BAI1-associated protein 2 (205294_at), score: -0.75 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: -0.92 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.9 CENPTcentromere protein T (218148_at), score: -0.78 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: -0.77 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.78 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.79 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: -0.78 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.96 FOXK2forkhead box K2 (203064_s_at), score: -0.89 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: -0.86 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: 0.88 IDUAiduronidase, alpha-L- (205059_s_at), score: -0.84 LOC339047hypothetical protein LOC339047 (221501_x_at), score: -0.79 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -1 MAP7D1MAP7 domain containing 1 (217943_s_at), score: -0.8 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: -0.9 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: -0.75 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: -0.87 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: -0.81 NPIPnuclear pore complex interacting protein (204538_x_at), score: -0.76 PARVBparvin, beta (37965_at), score: -0.75 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: -0.86 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: -0.92 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: -0.75 POLSpolymerase (DNA directed) sigma (202466_at), score: -0.85 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.85 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: -0.79 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: -0.83 RNF220ring finger protein 220 (219988_s_at), score: -0.86 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.94 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.92 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.94 SF1splicing factor 1 (208313_s_at), score: -0.84 SNAP23synaptosomal-associated protein, 23kDa (214544_s_at), score: 0.83 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.93 STRN4striatin, calmodulin binding protein 4 (217903_at), score: -0.87 TAPBPTAP binding protein (tapasin) (208829_at), score: -0.86 TXLNAtaxilin alpha (212300_at), score: -0.8 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.8 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: -0.79 ZNF536zinc finger protein 536 (206403_at), score: -0.76
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |