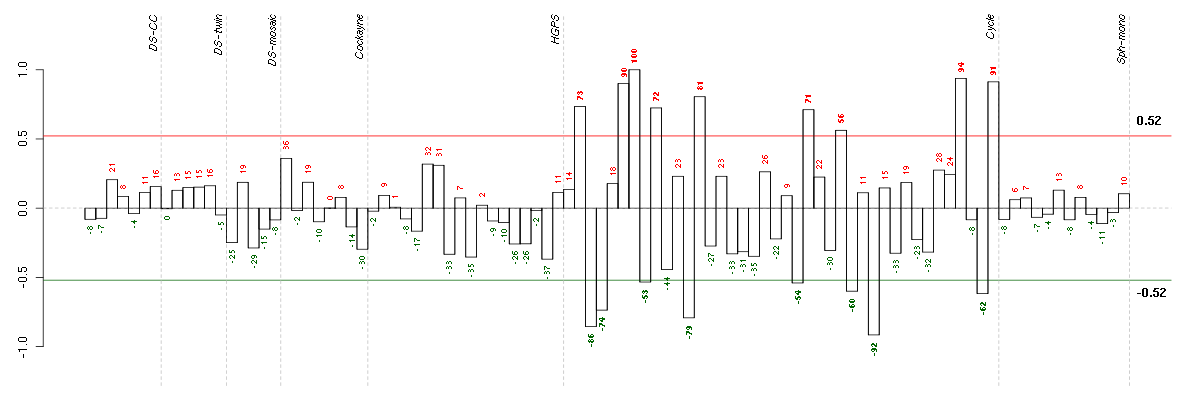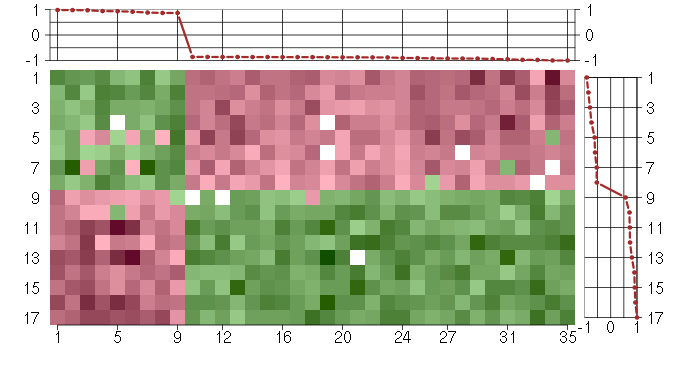

Under-expression is coded with green,
over-expression with red color.



ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.88 BAT2HLA-B associated transcript 2 (212081_x_at), score: -0.87 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.87 CALB2calbindin 2 (205428_s_at), score: 0.94 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.92 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: -0.86 CICcapicua homolog (Drosophila) (212784_at), score: -0.87 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.94 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.87 EPHA4EPH receptor A4 (206114_at), score: 0.87 FOXK2forkhead box K2 (203064_s_at), score: -0.98 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.91 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.98 JUPjunction plakoglobin (201015_s_at), score: -0.86 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.93 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.86 NDPNorrie disease (pseudoglioma) (206022_at), score: -0.95 NEFMneurofilament, medium polypeptide (205113_at), score: 0.97 NF1neurofibromin 1 (211094_s_at), score: -0.92 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: -0.87 OLFML2Bolfactomedin-like 2B (213125_at), score: -1 PCDH9protocadherin 9 (219737_s_at), score: 0.91 PXNpaxillin (211823_s_at), score: -0.97 RNF220ring finger protein 220 (219988_s_at), score: -0.87 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.98 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.88 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -1 SF1splicing factor 1 (208313_s_at), score: -0.87 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: -0.88 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.92 UBTD1ubiquitin domain containing 1 (219172_at), score: -0.86 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.92 WDR6WD repeat domain 6 (217734_s_at), score: -0.9 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: -0.88
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |