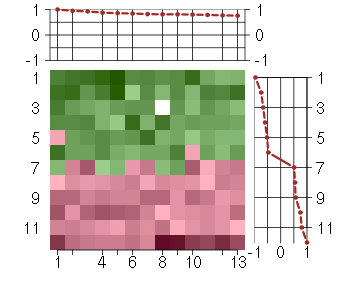

Under-expression is coded with green,
over-expression with red color.



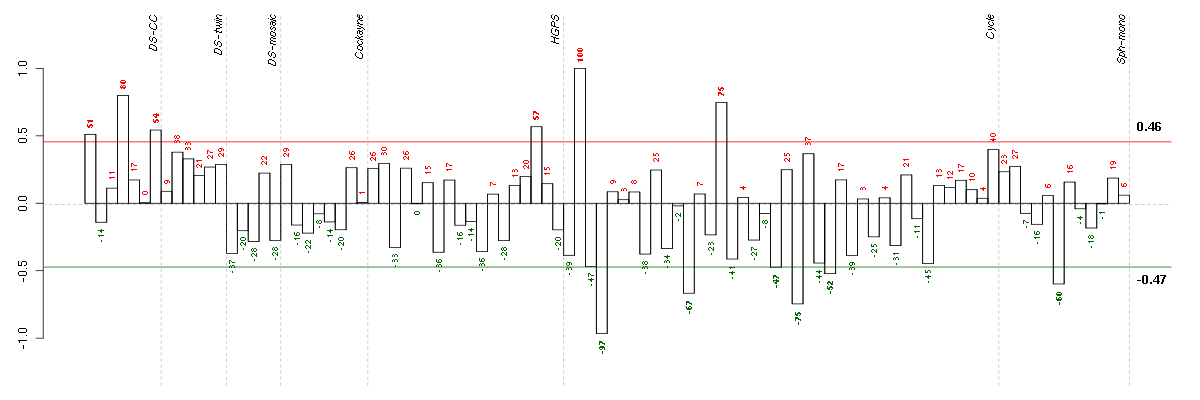
ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: 0.77 ADORA1adenosine A1 receptor (216220_s_at), score: 0.84 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: 0.81 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 1 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.95 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: 0.8 HAB1B1 for mucin (215778_x_at), score: 0.93 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: 0.81 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.88 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.82 LOC80054hypothetical LOC80054 (220465_at), score: 0.76 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.79 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.86
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |