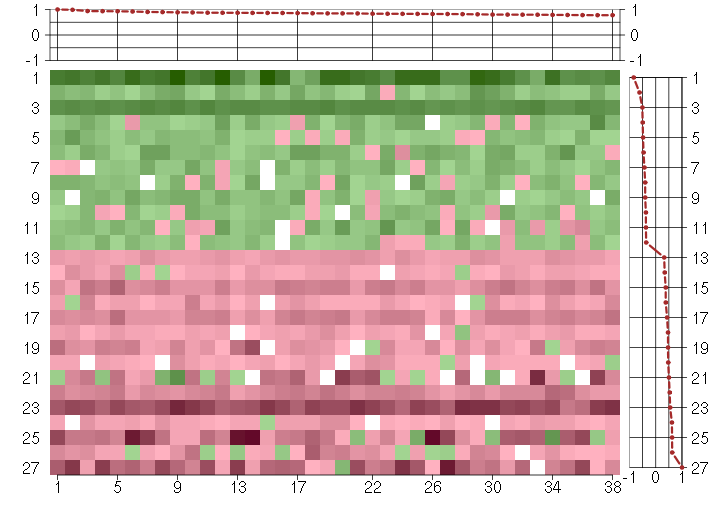

Under-expression is coded with green,
over-expression with red color.



ACOT11acyl-CoA thioesterase 11 (216103_at), score: 0.82 ADAMDEC1ADAM-like, decysin 1 (206134_at), score: 0.93 ADAMTS9ADAM metallopeptidase with thrombospondin type 1 motif, 9 (220287_at), score: 0.83 AFF2AF4/FMR2 family, member 2 (206105_at), score: 0.78 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: 0.85 CHN2chimerin (chimaerin) 2 (207486_x_at), score: 0.9 CLDN10claudin 10 (205328_at), score: 0.88 CTTNBP2NLCTTNBP2 N-terminal like (214731_at), score: 0.79 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (206919_at), score: 0.87 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (214053_at), score: 0.78 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: 1 KRT2keratin 2 (207908_at), score: 0.83 LAX1lymphocyte transmembrane adaptor 1 (207734_at), score: 0.83 LHX3LIM homeobox 3 (221670_s_at), score: 0.81 LOC100132247similar to Uncharacterized protein KIAA0220 (215002_at), score: 0.82 LOC93432maltase-glucoamylase-like pseudogene (216666_at), score: 0.85 MAP3K9mitogen-activated protein kinase kinase kinase 9 (213927_at), score: 0.87 MBmyoglobin (204179_at), score: 0.79 MCF2L2MCF.2 cell line derived transforming sequence-like 2 (215112_x_at), score: 0.78 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: 0.94 MYBPC1myosin binding protein C, slow type (214087_s_at), score: 0.82 MYO5Cmyosin VC (218966_at), score: 0.83 N4BP3Nedd4 binding protein 3 (214775_at), score: 0.8 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: 0.8 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: 0.87 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: 0.85 PEG3paternally expressed 3 (209242_at), score: 0.93 PROL1proline rich, lacrimal 1 (208004_at), score: 0.86 RP11-35N6.1plasticity related gene 3 (219732_at), score: 0.89 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: 0.85 SCARF1scavenger receptor class F, member 1 (206995_x_at), score: 0.92 SFTPBsurfactant protein B (214354_x_at), score: 0.9 SIRPB1signal-regulatory protein beta 1 (206934_at), score: 0.8 SLC12A3solute carrier family 12 (sodium/chloride transporters), member 3 (215274_at), score: 0.8 SLC6A14solute carrier family 6 (amino acid transporter), member 14 (219795_at), score: 0.87 SPAM1sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding) (216989_at), score: 0.86 TAAR2trace amine associated receptor 2 (221394_at), score: 0.99 TP63tumor protein p63 (209863_s_at), score: 0.87
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485731.cel | 5 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |