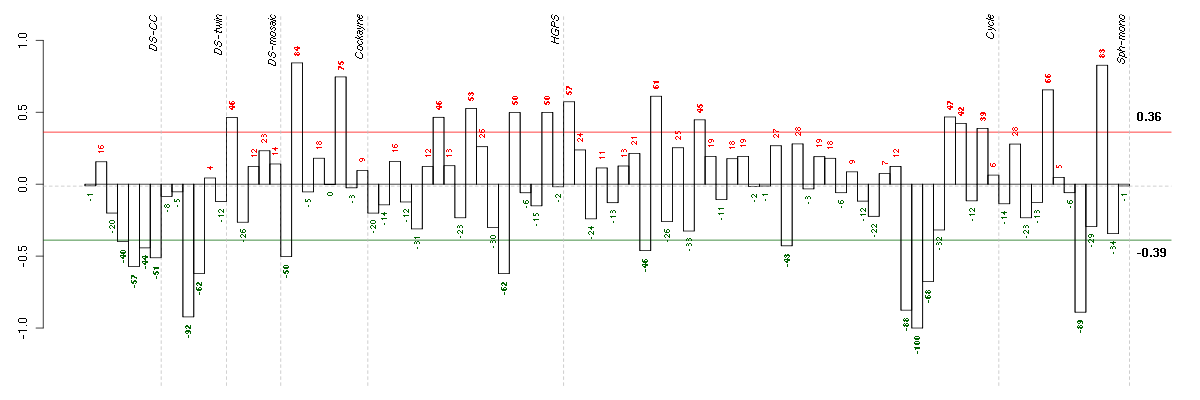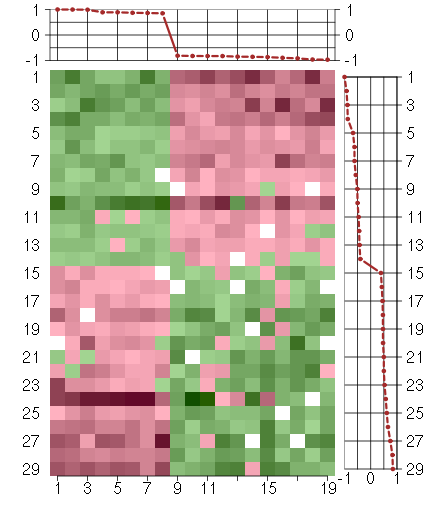

Under-expression is coded with green,
over-expression with red color.



BNIP3LBCL2/adenovirus E1B 19kDa interacting protein 3-like (221479_s_at), score: -0.83 C5orf4chromosome 5 open reading frame 4 (48031_r_at), score: -0.97 CD3EAPCD3e molecule, epsilon associated protein (205264_at), score: 1 CH25Hcholesterol 25-hydroxylase (206932_at), score: -0.89 DKC1dyskeratosis congenita 1, dyskerin (201478_s_at), score: 0.89 DTX4deltex homolog 4 (Drosophila) (212611_at), score: -0.82 FAM110Bfamily with sequence similarity 110, member B (221959_at), score: -0.83 FKBP4FK506 binding protein 4, 59kDa (200894_s_at), score: 0.85 HBP1HMG-box transcription factor 1 (209102_s_at), score: -0.92 KLHDC2kelch domain containing 2 (217906_at), score: -0.81 NIP7nuclear import 7 homolog (S. cerevisiae) (219031_s_at), score: 0.89 NOP16NOP16 nucleolar protein homolog (yeast) (203023_at), score: 0.87 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: -0.96 PPATphosphoribosyl pyrophosphate amidotransferase (209433_s_at), score: 1 QTRTD1queuine tRNA-ribosyltransferase domain containing 1 (219178_at), score: 0.86 RP11-345P4.4similar to solute carrier family 35, member E2 (217122_s_at), score: -0.85 SPRY1sprouty homolog 1, antagonist of FGF signaling (Drosophila) (212558_at), score: -0.86 UCK2uridine-cytidine kinase 2 (209825_s_at), score: 0.99 YPEL5yippee-like 5 (Drosophila) (217783_s_at), score: -0.87
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690376.cel | 13 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690392.cel | 14 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690472.cel | 17 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |