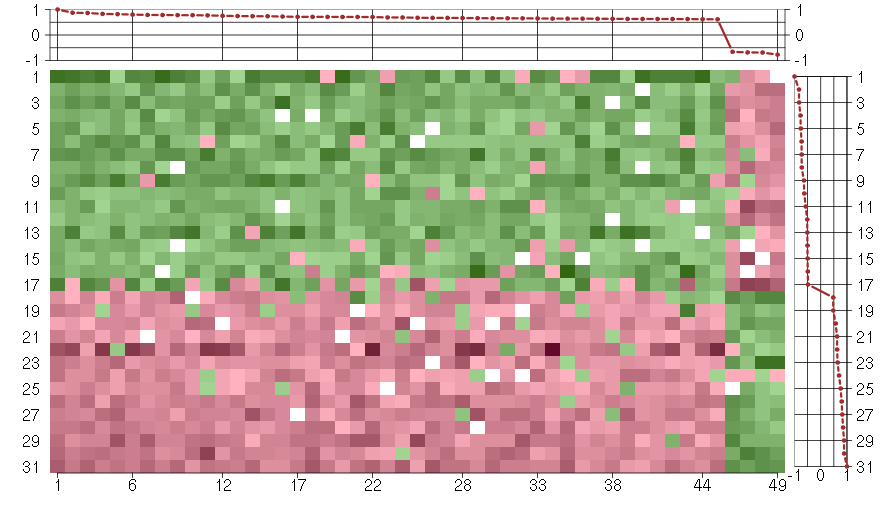

Under-expression is coded with green,
over-expression with red color.

biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of developmental process
Any process that modulates the frequency, rate or extent of development, the biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
negative regulation of developmental process
Any process that stops, prevents or reduces the rate or extent of development, the biological process whose specific outcome is the progression of an organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
negative regulation of developmental process
Any process that stops, prevents or reduces the rate or extent of development, the biological process whose specific outcome is the progression of an organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of developmental process
Any process that modulates the frequency, rate or extent of development, the biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
negative regulation of developmental process
Any process that stops, prevents or reduces the rate or extent of development, the biological process whose specific outcome is the progression of an organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).


| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 05220 | 1.626e-03 | 0.3333 | 5 | 64 | Chronic myeloid leukemia |
| 04010 | 1.428e-02 | 0.9531 | 6 | 183 | MAPK signaling pathway |
| 05212 | 2.026e-02 | 0.3438 | 4 | 66 | Pancreatic cancer |
| 04510 | 3.956e-02 | 0.7865 | 5 | 151 | Focal adhesion |
ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.68 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 0.78 ATF5activating transcription factor 5 (204999_s_at), score: 0.7 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.67 BCL2L1BCL2-like 1 (215037_s_at), score: 0.78 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.78 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: 0.67 C20orf27chromosome 20 open reading frame 27 (50314_i_at), score: 0.64 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.74 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.87 COL6A1collagen, type VI, alpha 1 (212940_at), score: 0.72 DENND3DENN/MADD domain containing 3 (212974_at), score: 0.67 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: 0.66 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: 0.65 FKRPfukutin related protein (219853_at), score: 0.63 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.66 FOXK2forkhead box K2 (203064_s_at), score: 0.69 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.86 LOC391132similar to hCG2041276 (216177_at), score: -0.68 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.65 MAP2K2mitogen-activated protein kinase kinase 2 (213490_s_at), score: 0.74 MUC1mucin 1, cell surface associated (207847_s_at), score: 0.63 NAB2NGFI-A binding protein 2 (EGR1 binding protein 2) (216017_s_at), score: 0.62 NF1neurofibromin 1 (211094_s_at), score: 0.77 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.74 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.62 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.8 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.83 PLEC1plectin 1, intermediate filament binding protein 500kDa (216971_s_at), score: 0.63 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.75 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.64 PXNpaxillin (211823_s_at), score: 1 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.69 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: 0.71 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.77 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.69 RXRBretinoid X receptor, beta (215099_s_at), score: 0.63 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.71 SENP3SUMO1/sentrin/SMT3 specific peptidase 3 (215113_s_at), score: 0.64 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.71 SHC1SHC (Src homology 2 domain containing) transforming protein 1 (201469_s_at), score: 0.63 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.63 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.79 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.71 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.71 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.66 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.82 WDR42AWD repeat domain 42A (202249_s_at), score: 0.64
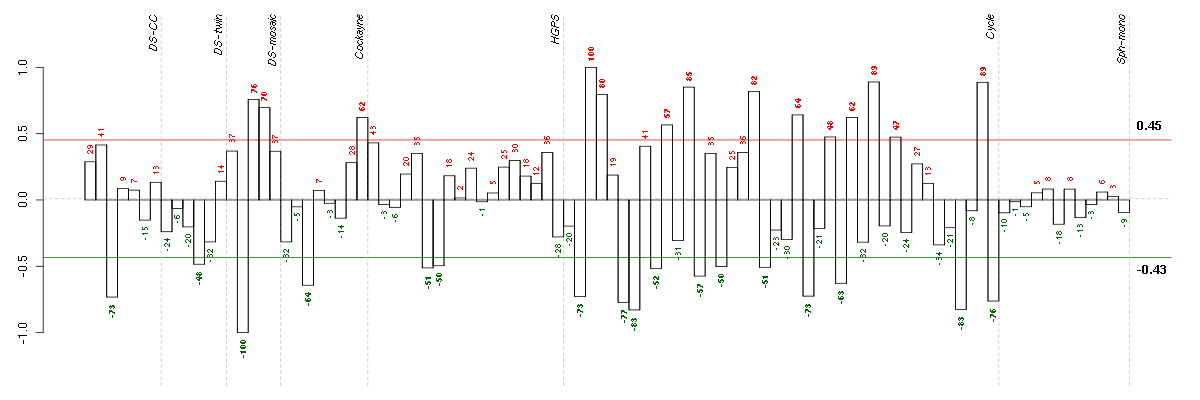
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690256.cel | 6 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |