Previous module |
Next module
Module #680, TG: 2.8, TC: 1.8, 98 probes, 97 Entrez genes, 5 conditions


HELP
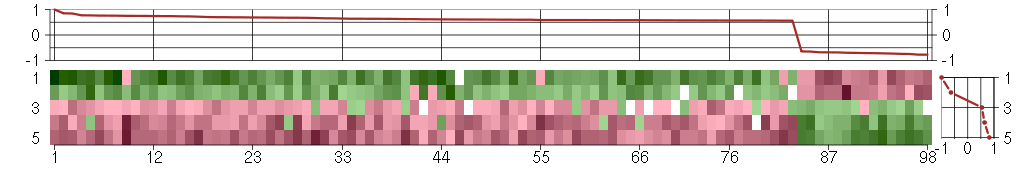
The image plot shows the color-coded level of gene expression, for the
genes and conditions in a given transcription module. The genes are on
the horizontal, the conditions on the vertical axis.
The genes are ordered according to their ISA gene scores, similarly
the conditions are ordered according to their condition scores. The
score of a gene means the «degree of inclusion» in
the module: a high score gene is essential in the module.
Condition scores can also be negative, that means that the genes of
the module are all down-regulated in the condition. Here the absolute
value of the score gives the «degree of inclusion».
The plots above and beside the expression matrix show the gene scores
and condition scores, respectively.
Note that the plot is interactive, you can see the name of the gene
and condition under the mouse cursor.
The expression matrix was normalized to have mean zero and standard
deviation one for every gene separately across all conditions
(i.e. not just for the conditions in the module).
— Click on the Help button again to close this help window.

Under-expression is coded with green,
over-expression with red color.
Help |
Hide |
Top
Help |
Show |
Top
The GO tree — Biological processes
HELP
This is one of three sections showing Gene Ontology enrichment of the
current module: in this case for biological processes.
The graph shows the hierarchy of the GO categories, their enrichment
for the current module is color coded, and the blue number beside the
category is the minus log ten p-value of the enrichment. (Calculated
using the standard hypergeometric test.) The color of the arrows code
«is a» (cyan) and «part of» relationships.
The tree was built the following way. First all GO terms with more
significant enrichment p-value than 0.05 were collected. Then all
paths from these terms to the root node of the GO tree were included
too. If a GO term is included more than once in the tree, then the
green numbers show 1) the id of the node, this makes it easier to find
other appereances of the term, and 2) the number of appearences.
Note that the same GO category might show up on the graph many
times. This is because the GO was «straightened» for this
graph, i.e. if there are more paths from a GO term to the root node of
the tree, all of them are included. The green numbers
Move the mouse cursor over the terms to get their definition. Clicking
on them takes you to the corresponding Gene Ontology web page.
If you cannot see a graph here at all, that means that there were no
significantly enriched GO categories, at the 0.05 level.
— Click on the Help button again to close this help window.

Help |
Hide |
Top
Help |
Show |
Top
The GO tree — Cellular Components
HELP
This is one of three sections showing Gene Ontology enrichment of the
current module: in this case for cellular components.
The graph shows the hierarchy of the GO categories, their enrichment
for the current module is color coded, and the blue number beside the
category is the minus log ten p-value of the enrichment. (Calculated
using the standard hypergeometric test.) The color of the arrows code
«is a» (cyan) and «part of» relationships.
The tree was built the following way. First all GO terms with more
significant enrichment p-value than 0.05 were collected. Then all
paths from these terms to the root node of the GO tree were included
too. If a GO term is included more than once in the tree, then the
green numbers show 1) the id of the node, this makes it easier to find
other appereances of the term, and 2) the number of appearences.
Note that the same GO category might show up on the graph many
times. This is because the GO was «straightened» for this
graph, i.e. if there are more paths from a GO term to the root node of
the tree, all of them are included. The green numbers
Move the mouse cursor over the terms to get their definition. Clicking
on them takes you to the corresponding Gene Ontology web page.
If you cannot see a graph here at all, that means that there were no
significantly enriched GO categories, at the 0.05 level.
— Click on the Help button again to close this help window.

Help |
Hide |
Top
Help |
Show |
Top
The GO tree — Molecular Function
HELP
This is one of three sections showing Gene Ontology enrichment of the
current module: in this case for molecular function.
The graph shows the hierarchy of the GO categories, their enrichment
for the current module is color coded, and the blue number beside the
category is the minus log ten p-value of the enrichment. (Calculated
using the standard hypergeometric test.) The color of the arrows code
«is a» (cyan) and «part of» relationships.
The tree was built the following way. First all GO terms with more
significant enrichment p-value than 0.05 were collected. Then all
paths from these terms to the root node of the GO tree were included
too. If a GO term is included more than once in the tree, then the
green numbers show 1) the id of the node, this makes it easier to find
other appereances of the term, and 2) the number of appearences.
Note that the same GO category might show up on the graph many
times. This is because the GO was «straightened» for this
graph, i.e. if there are more paths from a GO term to the root node of
the tree, all of them are included. The green numbers
Move the mouse cursor over the terms to get their definition. Clicking
on them takes you to the corresponding Gene Ontology web page.
If you cannot see a graph here at all, that means that there were no
significantly enriched GO categories, at the 0.05 level.
— Click on the Help button again to close this help window.

Help |
Hide |
Top
Help |
Show |
Top
GO BP test for over-representation
HELP
List of all enriched GO categories (biological processes), at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module annotated with the given GO term, just by chance.
- Count
is the number of genes in the module annotated with the given GO
term.
- Size is the total number of genes (in our universe)
annotated with the GO term.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
GO category.
Clicking on the GO identifiers takes you to the Gene Ontology web
pages.
— Click on the Help button again to close this help window.
| Id | Pvalue | ExpCount | Count | Size | Term |
| GO:0043062 | 1.943e-02 | 0.5732 | 5
ACAN, AGT, COL11A2, MUC5AC, POU4F1 | 55 | extracellular structure organization |
| GO:0046622 | 3.765e-02 | 0.04169 | 2
AGT, SASH3 | 4 | positive regulation of organ growth |
| GO:0030198 | 4.735e-02 | 0.4377 | 4
ACAN, AGT, COL11A2, MUC5AC | 42 | extracellular matrix organization |
| GO:0048704 | 4.850e-02 | 0.198 | 3
HOXA6, HOXB7, TGFBR2 | 19 | embryonic skeletal system morphogenesis |
Help |
Hide |
Top
Help |
Show |
Top
GO CC test for over-representation
HELP
List of all enriched GO categories (cellular components), at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module annotated with the given GO term, just by chance.
- Count
is the number of genes in the module annotated with the given GO
term.
- Size is the total number of genes (in our universe)
annotated with the GO term.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
GO category.
Clicking on the GO identifiers takes you to the Gene Ontology web
pages.
— Click on the Help button again to close this help window.
| Id | Pvalue | ExpCount | Count | Size | Term |
| GO:0005578 | 2.822e-02 | 1.606 | 7
ACAN, ADAMTS12, COL11A2, MMP17, MUC5AC, NYX, WNT6 | 153 | proteinaceous extracellular matrix |
| GO:0005576 | 3.607e-02 | 7.062 | 16
ACAN, ADAMTS12, AGT, COL11A2, FGL1, FLJ21075, GDF9, HAMP, LILRA5, MMP17, MUC13, MUC5AC, NYX, REG3A, ST6GAL1, WNT6 | 673 | extracellular region |
| GO:0031012 | 3.776e-02 | 1.711 | 7
ACAN, ADAMTS12, COL11A2, MMP17, MUC5AC, NYX, WNT6 | 163 | extracellular matrix |
| GO:0044421 | 4.220e-02 | 3.946 | 11
ACAN, ADAMTS12, AGT, COL11A2, FGL1, GDF9, MMP17, MUC5AC, NYX, REG3A, WNT6 | 376 | extracellular region part |
Help |
Hide |
Top
Help |
Show |
Top
GO MF test for over-representation
HELP
List of all enriched GO categories (molecular function), at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module annotated with the given GO term, just by chance.
- Count
is the number of genes in the module annotated with the given GO
term.
- Size is the total number of genes (in our universe)
annotated with the GO term.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
GO category.
Clicking on the GO identifiers takes you to the Gene Ontology web
pages.
— Click on the Help button again to close this help window.
| Id | Pvalue | ExpCount | Count | Size | Term |
| GO:0004871 | 3.669e-02 | 7.671 | 18
ABCC9, CD79B, CORIN, EPAG, FZD4, GPR116, GPR144, GPR32, ITGA6, ITGAL, LILRA5, MAPK10, OPRL1, PLK2, SIM2, TGFBR2, THRA, WNT6 | 780 | signal transducer activity |
| GO:0060089 | 3.669e-02 | 7.671 | 18
ABCC9, CD79B, CORIN, EPAG, FZD4, GPR116, GPR144, GPR32, ITGA6, ITGAL, LILRA5, MAPK10, OPRL1, PLK2, SIM2, TGFBR2, THRA, WNT6 | 780 | molecular transducer activity |
Help |
Hide |
Top
Help |
Show |
Top
KEGG Pathway test for over-representation
HELP
List of all enriched KEGG pathways, at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module annotated with the given KEGG pathway, just by chance.
- Count
is the number of genes in the module annotated with the given KEGG
pathway.
- Size is the total number of genes (in our universe)
annotated with the KEGG pathway.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
KEGG pathway.
Clicking on the KEGG identifiers takes you to the KEGG web site.
— Click on the Help button again to close this help window.
Help |
Hide |
Top
Help |
Show |
Top
miRNA test for over-representation
HELP
List of all enriched miRNA families, at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module regulated by the given miRNA family, just by chance.
- Count
is the number of genes in the module regulated by the given miRNA
family.
- Size is the total number of genes (in our universe)
regulated with the given miRNA family.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
miRNA family.
The miRNA regulation data was taken from the TargetScan database.
(Only the conserved sites were used for the current analysis.)
Clicking on the miRNA names takes you to the TargetScan web site.
— Click on the Help button again to close this help window.
Help |
Hide |
Top
Help |
Show |
Top
Chromosome test for over-representation
HELP
List of all enriched Chromosomes, at the 0.05
p-value level.
The columns:
- ExpCount is the expected number of genes in the
module on the given chromosome, just by chance.
- Count
is the number of genes in the module on the given chromosome.
- Size is the total number of genes (in our universe)
on the given chromosome.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
chromosome.
— Click on the Help button again to close this help window.
HELP
A list of all genes in the current module, in alphabetical order. The
size of the text corresponds to the gene scores.
Note that some gene symbols may show up more than once, if many
probes match the same Entrez gene.
Genes with no Entrez mapping are given separately, with their
Affymetrics probe ID.
— Click on the Help button again to close this help window.
Entrez genes
ABCC9ATP-binding cassette, sub-family C (CFTR/MRP), member 9 (208561_at), score: -0.68
ABCG4ATP-binding cassette, sub-family G (WHITE), member 4 (207593_at), score: 0.58
ACANaggrecan (205679_x_at), score: 0.68
ADAMTS12ADAM metallopeptidase with thrombospondin type 1 motif, 12 (221421_s_at), score: 0.57
AGTangiotensinogen (serpin peptidase inhibitor, clade A, member 8) (202834_at), score: 0.84
ALDH4A1aldehyde dehydrogenase 4 family, member A1 (211552_s_at), score: 0.64
ALPPL2alkaline phosphatase, placental-like 2 (216377_x_at), score: 0.57
ARHGEF10LRho guanine nucleotide exchange factor (GEF) 10-like (221656_s_at), score: 0.6
ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: 0.57
ATF3activating transcription factor 3 (202672_s_at), score: 0.61
C7orf58chromosome 7 open reading frame 58 (220032_at), score: -0.68
CALB1calbindin 1, 28kDa (205626_s_at), score: 0.74
CAPGcapping protein (actin filament), gelsolin-like (201850_at), score: -0.64
CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.69
COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.62
CORINcorin, serine peptidase (220356_at), score: 0.59
CPNE3copine III (202118_s_at), score: -0.71
CXorf56chromosome X open reading frame 56 (219805_at), score: 0.57
CYFIP2cytoplasmic FMR1 interacting protein 2 (215785_s_at), score: 0.57
DPYSL3dihydropyrimidinase-like 3 (201431_s_at), score: -0.65
DUSP8dual specificity phosphatase 8 (206374_at), score: 0.68
DUSP9dual specificity phosphatase 9 (205777_at), score: 0.57
EFHD1EF-hand domain family, member D1 (209343_at), score: 0.56
ENPEPglutamyl aminopeptidase (aminopeptidase A) (204844_at), score: 0.76
EPAGearly lymphoid activation protein (217050_at), score: 0.63
ETV7ets variant 7 (221680_s_at), score: 0.73
FBP1fructose-1,6-bisphosphatase 1 (209696_at), score: 0.76
FGL1fibrinogen-like 1 (205305_at), score: 0.6
FLJ21075hypothetical protein FLJ21075 (221172_at), score: 0.62
FZD4frizzled homolog 4 (Drosophila) (218665_at), score: 0.57
GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: 0.77
GDF9growth differentiation factor 9 (221314_at), score: 0.57
GPR116G protein-coupled receptor 116 (212950_at), score: 0.75
GPR144G protein-coupled receptor 144 (216289_at), score: 0.74
GPR32G protein-coupled receptor 32 (221469_at), score: 1
HAB1B1 for mucin (215778_x_at), score: 0.77
HAMPhepcidin antimicrobial peptide (220491_at), score: 0.63
HOXA6homeobox A6 (208557_at), score: 0.69
HOXB7homeobox B7 (204779_s_at), score: -0.75
IDI2isopentenyl-diphosphate delta isomerase 2 (217631_at), score: 0.61
INE1inactivation escape 1 (non-protein coding) (207252_at), score: 0.64
ITGA6integrin, alpha 6 (215177_s_at), score: -0.7
ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.7
KALRNkalirin, RhoGEF kinase (206078_at), score: 0.72
KCND3potassium voltage-gated channel, Shal-related subfamily, member 3 (213832_at), score: 0.6
KIAA1659KIAA1659 protein (215674_at), score: 0.74
LILRA5leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 (215838_at), score: 0.6
LOC149478hypothetical protein LOC149478 (215462_at), score: 0.75
LOH3CR2Aloss of heterozygosity, 3, chromosomal region 2, gene A (220244_at), score: 0.63
MAFGv-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) (204970_s_at), score: 0.73
MAP2microtubule-associated protein 2 (210015_s_at), score: 0.58
MAPK10mitogen-activated protein kinase 10 (204813_at), score: 0.58
MEGF8multiple EGF-like-domains 8 (214778_at), score: 0.57
MMP17matrix metallopeptidase 17 (membrane-inserted) (206234_s_at), score: 0.68
MOGmyelin oligodendrocyte glycoprotein (214650_x_at), score: 0.61
MTMR11myotubularin related protein 11 (205076_s_at), score: -0.68
MUC13mucin 13, cell surface associated (218687_s_at), score: 0.64
MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.59
N4BP3Nedd4 binding protein 3 (214775_at), score: 0.58
NACA2nascent polypeptide-associated complex alpha subunit 2 (222224_at), score: 0.61
NEFMneurofilament, medium polypeptide (205113_at), score: 0.68
NPEPL1aminopeptidase-like 1 (89476_r_at), score: 0.65
NYXnyctalopin (221684_s_at), score: 0.7
OPRL1opiate receptor-like 1 (206564_at), score: 0.59
PCNXpecanex homolog (Drosophila) (213159_at), score: -0.74
PDLIM3PDZ and LIM domain 3 (209621_s_at), score: 0.67
PHF7PHD finger protein 7 (215622_x_at), score: 0.85
PIP5K1Bphosphatidylinositol-4-phosphate 5-kinase, type I, beta (217477_at), score: 0.56
PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.6
PLK2polo-like kinase 2 (Drosophila) (201939_at), score: -0.72
PNMAL1PNMA-like 1 (218824_at), score: -0.73
POU4F1POU class 4 homeobox 1 (206940_s_at), score: 0.61
PPP4R4protein phosphatase 4, regulatory subunit 4 (220672_at), score: 0.58
PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.61
REG3Aregenerating islet-derived 3 alpha (205815_at), score: 0.65
SASH3SAM and SH3 domain containing 3 (204923_at), score: 0.58
SEC61A2Sec61 alpha 2 subunit (S. cerevisiae) (219499_at), score: -0.77
SENP7SUMO1/sentrin specific peptidase 7 (220735_s_at), score: -0.7
SETD1ASET domain containing 1A (213202_at), score: 0.57
SIM2single-minded homolog 2 (Drosophila) (206558_at), score: 0.62
SLC24A3solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 (57588_at), score: 0.56
SLC2A3solute carrier family 2 (facilitated glucose transporter), member 3 (202499_s_at), score: 0.69
SLC3A1solute carrier family 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1 (205799_s_at), score: 0.58
SPATA1spermatogenesis associated 1 (221057_at), score: 0.76
ST6GAL1ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (201998_at), score: 0.67
STYK1serine/threonine/tyrosine kinase 1 (221696_s_at), score: 0.58
SV2Asynaptic vesicle glycoprotein 2A (203069_at), score: 0.64
TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: -0.77
THRAthyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) (35846_at), score: 0.69
TMEM92transmembrane protein 92 (216791_at), score: 0.62
TSPAN15tetraspanin 15 (218693_at), score: 0.75
TTTY9Atestis-specific transcript, Y-linked 9A (211460_at), score: 0.59
TULP2tubby like protein 2 (206733_at), score: 0.58
TYMPthymidine phosphorylase (217497_at), score: 0.7
VAMP8vesicle-associated membrane protein 8 (endobrevin) (202546_at), score: 0.59
WNT6wingless-type MMTV integration site family, member 6 (71933_at), score: 0.58
ZNF185zinc finger protein 185 (LIM domain) (203585_at), score: -0.71
Non-Entrez genes
203069_atUnknown, score: 0.57
HELP
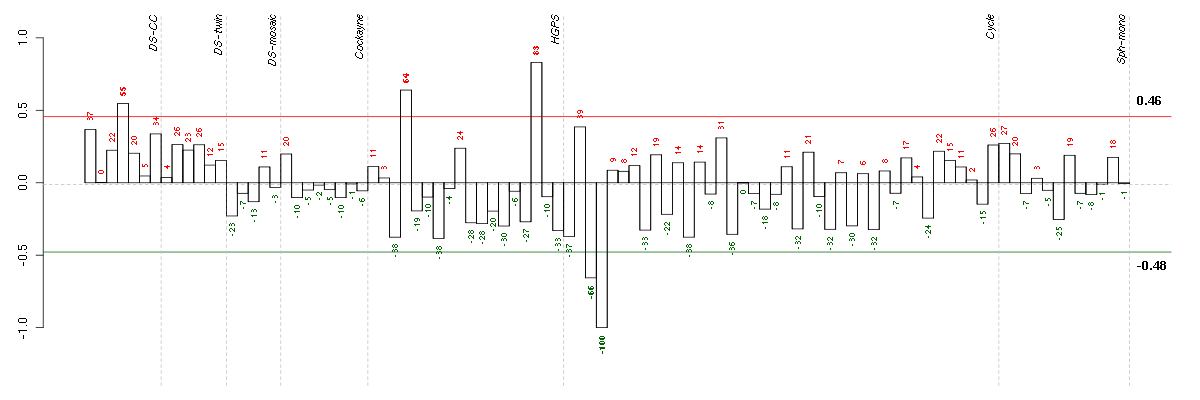
Conditions in the module, given in the same order as on the expression
plot above. Red color means over-expression, green under-expression in
the given condition.
The barplot below shows the condition (sample) scores. A separate bar
is shown for each sample, its height is the corresponding score of the
sample in the module. The red and green numbers on the bars are the
sample scores expressed in percents, i.e. 100% is 1.0.
The red and green lines show the module thresholds, samples above
the red line and below the green line are included in the module.
The different experiments that were part of the study, are separated
by dashed vertical lines.
— Click on the Help button again to close this help window.
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |



© 2008-2010 Computational Biology Group, Department of Medical Genetics,
University of Lausanne, Switzerland