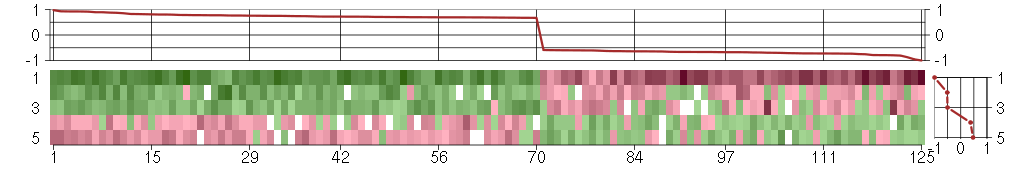

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.

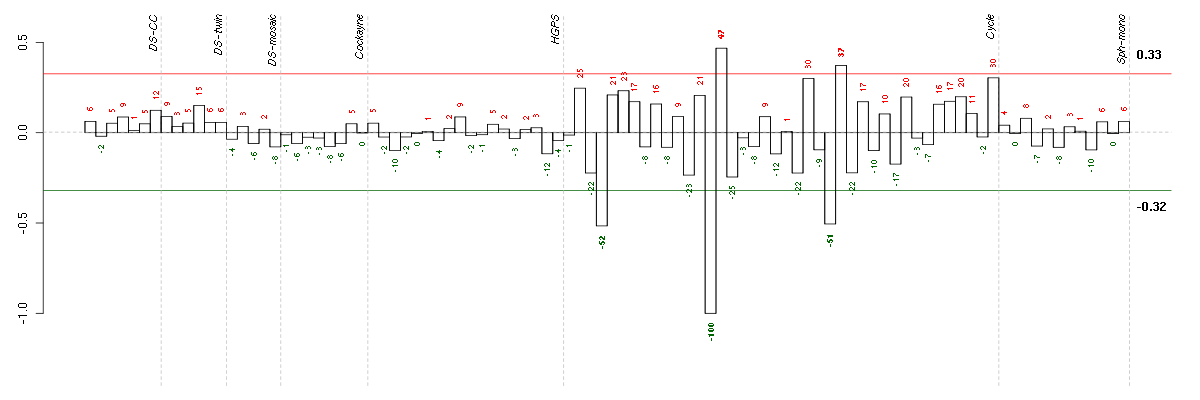
ACSL6acyl-CoA synthetase long-chain family member 6 (211207_s_at), score: -0.69 ACTN3actinin, alpha 3 (206891_at), score: 0.78 ADAMDEC1ADAM-like, decysin 1 (206134_at), score: -0.72 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.73 ALDH4A1aldehyde dehydrogenase 4 family, member A1 (211552_s_at), score: 0.75 ANGEL1angel homolog 1 (Drosophila) (36865_at), score: 0.69 APOC1apolipoprotein C-I (213553_x_at), score: 0.75 ASTN2astrotactin 2 (209693_at), score: 0.69 ATPAF2ATP synthase mitochondrial F1 complex assembly factor 2 (213057_at), score: 0.69 AVILadvillin (205539_at), score: 0.75 C1orf66chromosome 1 open reading frame 66 (218914_at), score: 0.69 C8orf44chromosome 8 open reading frame 44 (220216_at), score: 0.72 CCDC121coiled-coil domain containing 121 (220321_s_at), score: -0.67 CD200CD200 molecule (209583_s_at), score: -0.65 CD53CD53 molecule (203416_at), score: 0.75 CDH19cadherin 19, type 2 (206898_at), score: -0.66 CDRT1CMT1A duplicated region transcript 1 (215999_at), score: 0.7 CHRNA5cholinergic receptor, nicotinic, alpha 5 (206533_at), score: -0.66 CLTCL1clathrin, heavy chain-like 1 (205944_s_at), score: 0.69 CNR2cannabinoid receptor 2 (macrophage) (206586_at), score: 0.88 COL9A2collagen, type IX, alpha 2 (213622_at), score: 0.82 COX6A2cytochrome c oxidase subunit VIa polypeptide 2 (206353_at), score: 0.7 CPNE3copine III (202118_s_at), score: -0.63 CTSScathepsin S (202901_x_at), score: -0.96 CXCL11chemokine (C-X-C motif) ligand 11 (210163_at), score: -0.73 CYP26B1cytochrome P450, family 26, subfamily B, polypeptide 1 (219825_at), score: -0.73 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: 0.67 DSCR6Down syndrome critical region gene 6 (207267_s_at), score: -0.64 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.97 EGLN3egl nine homolog 3 (C. elegans) (219232_s_at), score: -0.61 ELLelongation factor RNA polymerase II (204095_s_at), score: 0.69 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: 0.71 EPHA3EPH receptor A3 (211164_at), score: -0.72 EPM2Aepilepsy, progressive myoclonus type 2A, Lafora disease (laforin) (205231_s_at), score: 0.82 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (214053_at), score: -0.75 F7coagulation factor VII (serum prothrombin conversion accelerator) (207300_s_at), score: 0.76 FBP1fructose-1,6-bisphosphatase 1 (209696_at), score: 0.76 FBRSfibrosin (218255_s_at), score: 0.92 FBXO2F-box protein 2 (219305_x_at), score: 0.81 FBXO22F-box protein 22 (219638_at), score: -0.64 FGL1fibrinogen-like 1 (205305_at), score: 0.76 FKRPfukutin related protein (219853_at), score: -0.7 FOXK2forkhead box K2 (203064_s_at), score: -0.69 FRMD4BFERM domain containing 4B (213056_at), score: -0.67 GGT1gamma-glutamyltransferase 1 (209919_x_at), score: 0.78 GGTLC1gamma-glutamyltransferase light chain 1 (211416_x_at), score: 0.73 GPR144G protein-coupled receptor 144 (216289_at), score: 0.72 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: -0.81 GSTCDglutathione S-transferase, C-terminal domain containing (220063_at), score: -0.87 HAB1B1 for mucin (215778_x_at), score: 0.92 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: 0.89 HCG8HLA complex group 8 (215985_at), score: -1 HIST1H2ALhistone cluster 1, H2al (214554_at), score: -0.7 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: 0.67 IGFALSinsulin-like growth factor binding protein, acid labile subunit (215712_s_at), score: 0.7 IGLL3immunoglobulin lambda-like polypeptide 3 (215946_x_at), score: 0.72 ITGA10integrin, alpha 10 (206766_at), score: 0.8 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.87 JAG2jagged 2 (32137_at), score: 0.7 JAK2Janus kinase 2 (a protein tyrosine kinase) (205842_s_at), score: -0.59 KIAA1107KIAA1107 (214098_at), score: -0.6 KIF26Bkinesin family member 26B (220002_at), score: 0.76 KLF12Kruppel-like factor 12 (208467_at), score: -0.67 KLF7Kruppel-like factor 7 (ubiquitous) (204334_at), score: 0.76 LAT2linker for activation of T cells family, member 2 (211768_at), score: 0.89 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: 0.93 LOC100132247similar to Uncharacterized protein KIAA0220 (215002_at), score: -0.76 LOC391132similar to hCG2041276 (216177_at), score: 0.74 LOC729806similar to hCG1725380 (217544_at), score: -0.64 LPAR4lysophosphatidic acid receptor 4 (206960_at), score: 0.68 MANBAmannosidase, beta A, lysosomal (203778_at), score: 0.79 MAP3K9mitogen-activated protein kinase kinase kinase 9 (213927_at), score: -0.72 MCM3APASMCM3AP antisense RNA (non-protein coding) (220459_at), score: -0.6 MGAT4Cmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) (207447_s_at), score: -0.8 MGC5590hypothetical protein MGC5590 (220931_at), score: -0.67 MYBPC1myosin binding protein C, slow type (214087_s_at), score: -0.66 NACA2nascent polypeptide-associated complex alpha subunit 2 (222224_at), score: 0.72 NEURLneuralized homolog (Drosophila) (204889_s_at), score: 0.72 NFYCnuclear transcription factor Y, gamma (202215_s_at), score: -0.64 NMUneuromedin U (206023_at), score: -0.67 NOTCH4Notch homolog 4 (Drosophila) (205247_at), score: 0.74 OR10C1olfactory receptor, family 10, subfamily C, member 1 (221339_at), score: -0.67 PARD6Apar-6 partitioning defective 6 homolog alpha (C. elegans) (205245_at), score: 0.74 PDE3Aphosphodiesterase 3A, cGMP-inhibited (206388_at), score: -0.79 PGCprogastricsin (pepsinogen C) (205261_at), score: 0.92 PIAS4protein inhibitor of activated STAT, 4 (212881_at), score: 0.7 PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.8 PLS1plastin 1 (I isoform) (205190_at), score: -0.67 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: -0.59 PRKXprotein kinase, X-linked (204061_at), score: -0.61 PRLHprolactin releasing hormone (221443_x_at), score: 0.7 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.77 PXNpaxillin (211823_s_at), score: -0.63 R3HDM2R3H domain containing 2 (203831_at), score: 0.78 RECQL5RecQ protein-like 5 (34063_at), score: -0.79 RNASE2ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) (206111_at), score: -0.72 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (220738_s_at), score: -0.68 SAA4serum amyloid A4, constitutive (207096_at), score: 0.82 SCN1Bsodium channel, voltage-gated, type I, beta (205508_at), score: 0.72 SEC14L5SEC14-like 5 (S. cerevisiae) (210169_at), score: 0.69 SFNstratifin (33322_i_at), score: 0.69 SH2D3ASH2 domain containing 3A (222169_x_at), score: 0.85 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: 0.92 SLC12A3solute carrier family 12 (sodium/chloride transporters), member 3 (215274_at), score: -0.64 SLC39A9solute carrier family 39 (zinc transporter), member 9 (217859_s_at), score: -0.6 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.72 SLC6A14solute carrier family 6 (amino acid transporter), member 14 (219795_at), score: -0.71 SPON1spondin 1, extracellular matrix protein (209436_at), score: -0.6 SPTA1spectrin, alpha, erythrocytic 1 (elliptocytosis 2) (206937_at), score: -0.67 ST6GAL1ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (201998_at), score: 0.7 TAAR2trace amine associated receptor 2 (221394_at), score: -0.79 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: 0.8 TLR1toll-like receptor 1 (210176_at), score: -0.63 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: 0.77 TNK1tyrosine kinase, non-receptor, 1 (217149_x_at), score: 0.68 TRAPPC9trafficking protein particle complex 9 (221836_s_at), score: 0.71 TSPAN9tetraspanin 9 (220968_s_at), score: 0.72 TWISTNBTWIST neighbor (214729_at), score: -0.7 VLDLRvery low density lipoprotein receptor (209822_s_at), score: 0.68 ZCCHC4zinc finger, CCHC domain containing 4 (220473_s_at), score: -0.62 ZCCHC6zinc finger, CCHC domain containing 6 (220933_s_at), score: 0.67 ZNF175zinc finger protein 175 (205497_at), score: -0.72 ZNF529zinc finger protein 529 (215307_at), score: 0.78 ZNF749zinc finger protein 749 (215289_at), score: -0.73 ZNF783zinc finger family member 783 (221876_at), score: 0.69
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |