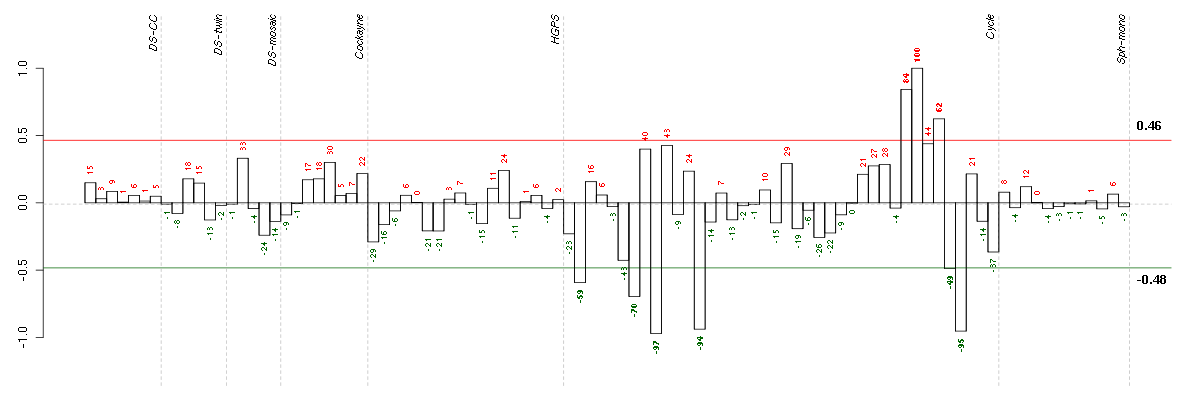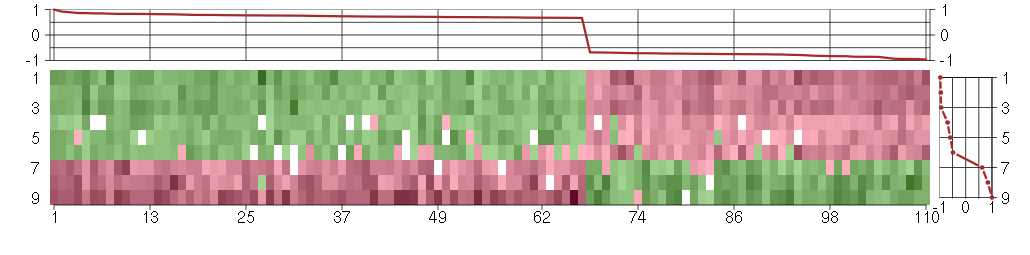

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.76 ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: 0.67 ADORA2Badenosine A2b receptor (205891_at), score: 0.84 AHI1Abelson helper integration site 1 (221569_at), score: -0.89 AIM1absent in melanoma 1 (212543_at), score: 0.79 ANGPTL2angiopoietin-like 2 (213004_at), score: 0.73 ANXA10annexin A10 (210143_at), score: -0.94 APOL6apolipoprotein L, 6 (219716_at), score: 0.85 ARHGEF2rho/rac guanine nucleotide exchange factor (GEF) 2 (209435_s_at), score: 0.76 ARL4CADP-ribosylation factor-like 4C (202207_at), score: 0.73 BMP6bone morphogenetic protein 6 (206176_at), score: -0.78 BTG2BTG family, member 2 (201236_s_at), score: 0.83 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: 0.89 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.78 CCDC28Acoiled-coil domain containing 28A (209479_at), score: 0.75 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: -0.73 CRBNcereblon (218142_s_at), score: 0.74 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.86 DAAM2dishevelled associated activator of morphogenesis 2 (212793_at), score: 0.68 DAPK1death-associated protein kinase 1 (203139_at), score: 0.83 DCLK1doublecortin-like kinase 1 (205399_at), score: 0.76 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.77 DEPDC6DEP domain containing 6 (218858_at), score: 0.92 DHRS3dehydrogenase/reductase (SDR family) member 3 (202481_at), score: 0.8 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.75 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.74 DOCK10dedicator of cytokinesis 10 (219279_at), score: -0.85 DUSP4dual specificity phosphatase 4 (204014_at), score: -0.74 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.76 ENOX1ecto-NOX disulfide-thiol exchanger 1 (219501_at), score: 0.81 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.82 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: -0.75 FGF9fibroblast growth factor 9 (glia-activating factor) (206404_at), score: 0.85 FNBP1formin binding protein 1 (212288_at), score: 0.69 GALK2galactokinase 2 (205219_s_at), score: -0.72 GDF15growth differentiation factor 15 (221577_x_at), score: 0.68 GDPD5glycerophosphodiester phosphodiesterase domain containing 5 (32502_at), score: 0.75 GKglycerol kinase (207387_s_at), score: -0.93 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.96 GRB14growth factor receptor-bound protein 14 (206204_at), score: -0.75 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 1 HSD17B7hydroxysteroid (17-beta) dehydrogenase 7 (220081_x_at), score: 0.77 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.71 IFIH1interferon induced with helicase C domain 1 (219209_at), score: 0.86 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.71 IL33interleukin 33 (209821_at), score: -0.86 INPP5Binositol polyphosphate-5-phosphatase, 75kDa (213804_at), score: 0.69 IRF1interferon regulatory factor 1 (202531_at), score: 0.72 JUNDjun D proto-oncogene (203751_x_at), score: 0.72 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.74 KIAA1305KIAA1305 (220911_s_at), score: 0.71 KIAA1462KIAA1462 (213316_at), score: 0.79 LDLRlow density lipoprotein receptor (202068_s_at), score: 0.73 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.83 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.82 LOC399491LOC399491 protein (214035_x_at), score: 0.79 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (211019_s_at), score: 0.71 LYPD1LY6/PLAUR domain containing 1 (212909_at), score: -0.8 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.77 MBPmyelin basic protein (210136_at), score: 0.72 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.73 MGC87042similar to Six transmembrane epithelial antigen of prostate (217553_at), score: -0.75 MKNK2MAP kinase interacting serine/threonine kinase 2 (218205_s_at), score: 0.7 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.76 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.76 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: -0.85 MX2myxovirus (influenza virus) resistance 2 (mouse) (204994_at), score: 0.72 NCALDneurocalcin delta (211685_s_at), score: -0.71 NEFMneurofilament, medium polypeptide (205113_at), score: -0.94 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.83 NPTX1neuronal pentraxin I (204684_at), score: -0.94 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.69 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.71 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: 0.82 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.69 PCDH9protocadherin 9 (219737_s_at), score: -0.86 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.7 PMP22peripheral myelin protein 22 (210139_s_at), score: 0.7 PPATphosphoribosyl pyrophosphate amidotransferase (209433_s_at), score: -0.73 PPLperiplakin (203407_at), score: 0.7 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: 0.78 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: 0.79 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.7 SELPLGselectin P ligand (209879_at), score: -0.68 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: -0.73 SIX2SIX homeobox 2 (206510_at), score: 0.76 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.75 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.83 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (207043_s_at), score: 0.72 SNNstannin (218032_at), score: 0.77 SSTR1somatostatin receptor 1 (208482_at), score: -0.81 SV2Asynaptic vesicle glycoprotein 2A (203069_at), score: 0.67 SYNPOsynaptopodin (202796_at), score: 0.67 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.68 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.81 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.7 TLR3toll-like receptor 3 (206271_at), score: 0.74 TM4SF1transmembrane 4 L six family member 1 (215034_s_at), score: -0.83 TMEM2transmembrane protein 2 (218113_at), score: -0.83 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.68 TNFSF9tumor necrosis factor (ligand) superfamily, member 9 (206907_at), score: 0.72 TP53tumor protein p53 (201746_at), score: 0.76 TRIM21tripartite motif-containing 21 (204804_at), score: 0.68 TSPAN13tetraspanin 13 (217979_at), score: -0.68 UHRF1BP1LUHRF1 binding protein 1-like (213118_at), score: -0.69 ULBP2UL16 binding protein 2 (221291_at), score: -0.78 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.7 URB1URB1 ribosome biogenesis 1 homolog (S. cerevisiae) (212996_s_at), score: -0.7 XAF1XIAP associated factor 1 (206133_at), score: 0.73 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.76
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |