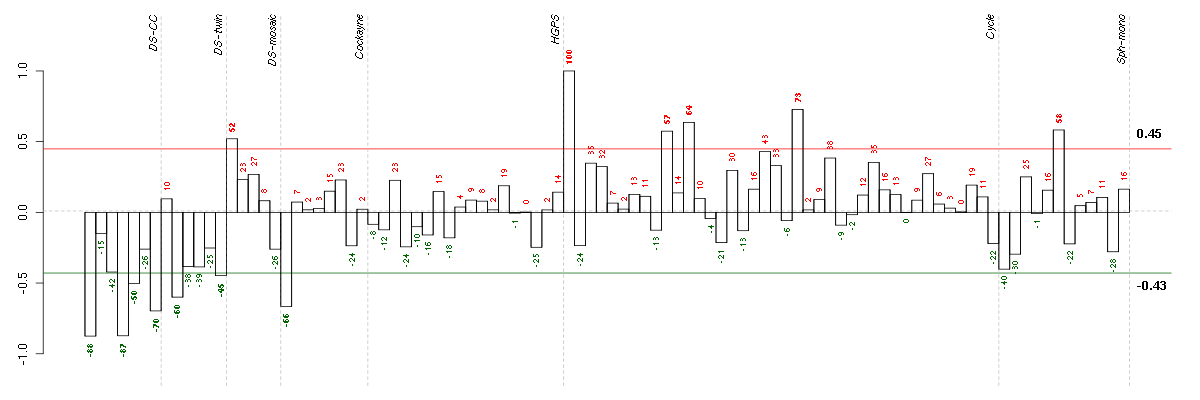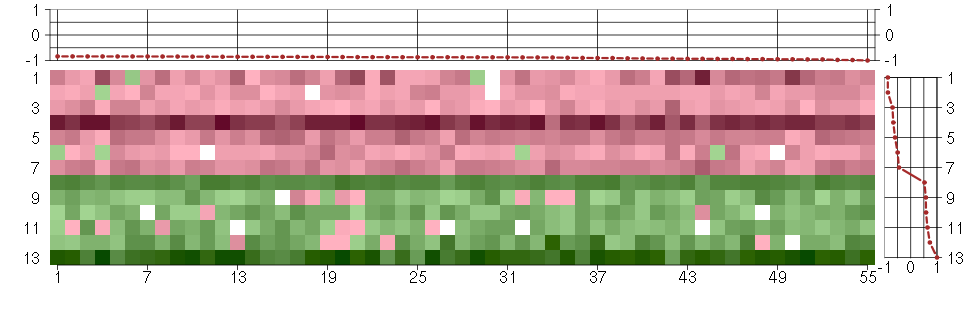

Under-expression is coded with green,
over-expression with red color.



A1CFAPOBEC1 complementation factor (220951_s_at), score: -0.94 ADAMTS9ADAM metallopeptidase with thrombospondin type 1 motif, 9 (220287_at), score: -0.84 AIPL1aryl hydrocarbon receptor interacting protein-like 1 (219977_at), score: -0.84 BMP10bone morphogenetic protein 10 (208292_at), score: -0.9 BTNL3butyrophilin-like 3 (217207_s_at), score: -0.92 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: -0.87 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: -0.87 C8orf60chromosome 8 open reading frame 60 (220712_at), score: -0.93 CEACAM5carcinoembryonic antigen-related cell adhesion molecule 5 (201884_at), score: -0.86 CEACAM6carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) (211657_at), score: -0.87 CHN2chimerin (chimaerin) 2 (207486_x_at), score: -0.85 CHST4carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 (220446_s_at), score: -0.87 CLDN10claudin 10 (205328_at), score: -0.94 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: -0.98 CTSEcathepsin E (205927_s_at), score: -0.88 CYP2A6cytochrome P450, family 2, subfamily A, polypeptide 6 (211295_x_at), score: -0.86 DNASE1L3deoxyribonuclease I-like 3 (205554_s_at), score: -0.88 DRD5dopamine receptor D5 (208486_at), score: -0.92 FAM38Bfamily with sequence similarity 38, member B (219602_s_at), score: -0.93 FOLH1folate hydrolase (prostate-specific membrane antigen) 1 (217487_x_at), score: -0.9 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: -0.87 KIR3DX1killer cell immunoglobulin-like receptor, three domains, X1 (216428_x_at), score: -0.86 KLF1Kruppel-like factor 1 (erythroid) (210504_at), score: -0.89 LHX3LIM homeobox 3 (221670_s_at), score: -0.84 LOC100188945cell division cycle associated 4 pseudogene (215109_at), score: -0.95 MMRN2multimerin 2 (219091_s_at), score: -0.87 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: -0.96 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.85 MYO1Amyosin IA (211916_s_at), score: -0.84 N4BP3Nedd4 binding protein 3 (214775_at), score: -0.86 NOTCH4Notch homolog 4 (Drosophila) (205247_at), score: -0.87 NRN1neuritin 1 (218625_at), score: -0.86 OR7E24olfactory receptor, family 7, subfamily E, member 24 (215463_at), score: -0.93 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: -0.96 PON1paraoxonase 1 (206344_at), score: -0.87 PPP4R4protein phosphatase 4, regulatory subunit 4 (220672_at), score: -0.84 PRB3proline-rich protein BstNI subfamily 3 (206998_x_at), score: -0.86 PRKCBprotein kinase C, beta (209685_s_at), score: -0.84 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: -0.95 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: -1 RETret proto-oncogene (205879_x_at), score: -0.85 ROS1c-ros oncogene 1 , receptor tyrosine kinase (207569_at), score: -0.84 RP11-35N6.1plasticity related gene 3 (219732_at), score: -0.85 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: -0.98 S100A14S100 calcium binding protein A14 (218677_at), score: -0.94 SERPINA1serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 (211429_s_at), score: -0.88 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: -0.87 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (211349_at), score: -0.9 SLC26A10solute carrier family 26, member 10 (214951_at), score: -0.88 TACR1tachykinin receptor 1 (208048_at), score: -0.85 TBX21T-box 21 (220684_at), score: -0.88 TBX6T-box 6 (207684_at), score: -0.84 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.85 TRA@T cell receptor alpha locus (216540_at), score: -0.95 VGFVGF nerve growth factor inducible (205586_x_at), score: -0.93
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |