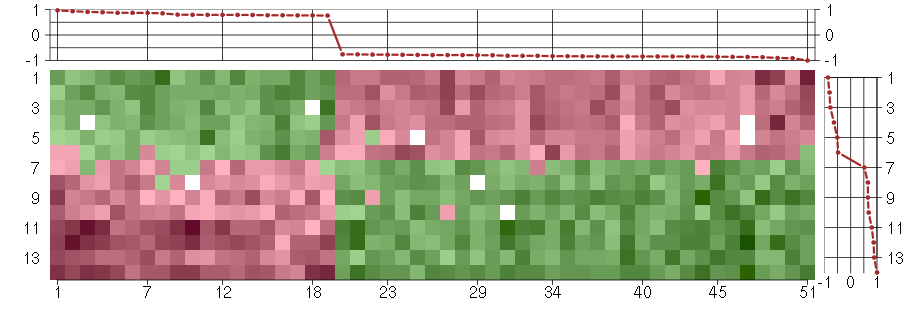

Under-expression is coded with green,
over-expression with red color.



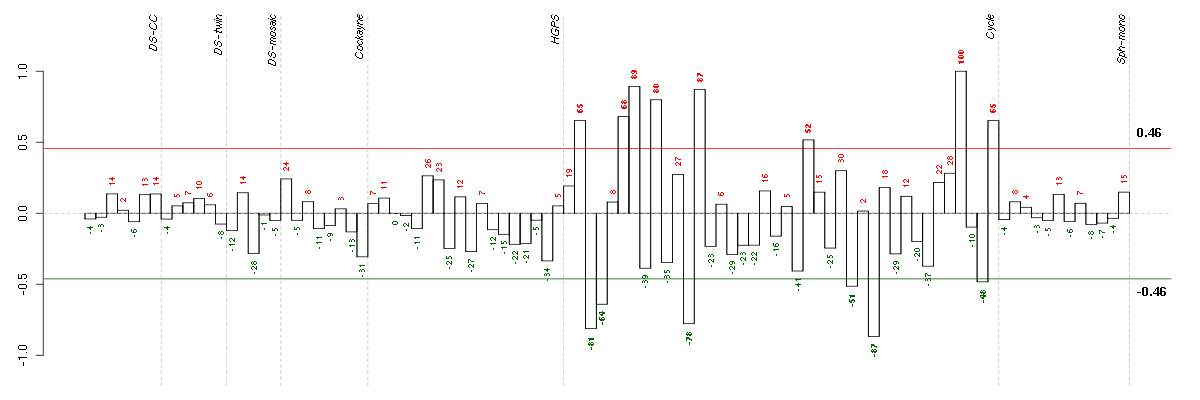
ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: 0.8 BAT2HLA-B associated transcript 2 (212081_x_at), score: -0.8 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.77 CALB2calbindin 2 (205428_s_at), score: 0.87 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.84 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: -0.83 CICcapicua homolog (Drosophila) (212784_at), score: -0.84 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: -0.76 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.84 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.86 DENND3DENN/MADD domain containing 3 (212974_at), score: -0.79 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: 0.89 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: -0.85 FOXK2forkhead box K2 (203064_s_at), score: -0.85 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: 0.76 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: 0.79 H1FXH1 histone family, member X (204805_s_at), score: -0.82 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.84 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.87 JUPjunction plakoglobin (201015_s_at), score: -0.84 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.91 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: 0.77 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: -0.77 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.78 NDPNorrie disease (pseudoglioma) (206022_at), score: -0.9 NEFMneurofilament, medium polypeptide (205113_at), score: 0.97 NF1neurofibromin 1 (211094_s_at), score: -0.87 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: -0.79 OLFML2Bolfactomedin-like 2B (213125_at), score: -1 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: 0.79 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: -0.79 PCDH9protocadherin 9 (219737_s_at), score: 0.93 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: -0.82 PRMT3protein arginine methyltransferase 3 (213320_at), score: 0.85 PXNpaxillin (211823_s_at), score: -0.84 RAB1ARAB1A, member RAS oncogene family (213440_at), score: 0.78 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: -0.84 RNF220ring finger protein 220 (219988_s_at), score: -0.79 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.87 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.77 SAP30LSAP30-like (219129_s_at), score: -0.76 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.91 SF1splicing factor 1 (208313_s_at), score: -0.84 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.78 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: 0.79 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.79 TLE4transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) (204872_at), score: 0.77 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.83 WDR6WD repeat domain 6 (217734_s_at), score: -0.86 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: -0.82
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |