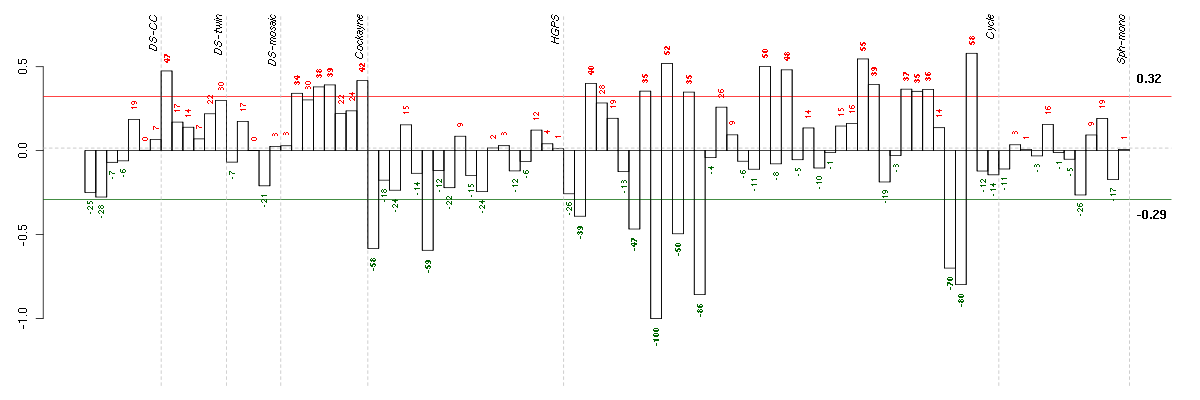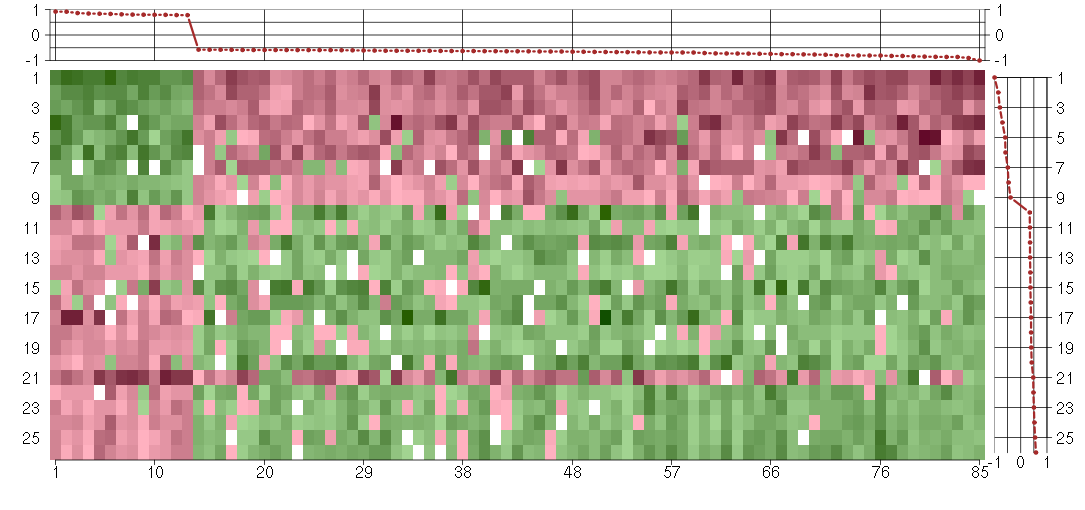

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of nucleotide metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleotides.
cAMP biosynthetic process
The chemical reactions and pathways resulting in the formation of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
nucleoside phosphate metabolic process
The chemical reactions and pathways involving any phosphorylated nucleoside.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cell proliferation
The multiplication or reproduction of cells, resulting in the expansion of a cell population.
cAMP metabolic process
The chemical reactions and pathways involving the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
nucleotide metabolic process
The chemical reactions and pathways involving a nucleotide, a nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the glycose moiety; may be mono-, di- or triphosphate; this definition includes cyclic nucleotides (nucleoside cyclic phosphates).
nucleotide biosynthetic process
The chemical reactions and pathways resulting in the formation of nucleotides, any nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the glycose moiety; may be mono-, di- or triphosphate; this definition includes cyclic-nucleotides (nucleoside cyclic phosphates).
cyclic nucleotide metabolic process
The chemical reactions and pathways involving a cyclic nucleotide, a nucleotide in which the phosphate group is in diester linkage to two positions on the sugar residue.
cyclic nucleotide biosynthetic process
The chemical reactions and pathways resulting in the formation of a cyclic nucleotide, a nucleotide in which the phosphate group is in diester linkage to two positions on the sugar residue.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cyclic nucleotide metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving cyclic nucleotides.
regulation of cyclic nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of cyclic nucleotides.
regulation of nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of nucleotides.
regulation of cAMP metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
regulation of cAMP biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
regulation of cyclase activity
Any process that modulates the frequency, rate or extent of cyclase activity.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of cell proliferation
Any process that modulates the frequency, rate or extent of cell proliferation.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of adenylate cyclase activity
Any process that modulates the frequency, rate or extent of adenylate cyclase activity.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of lyase activity
Any process that modulates the frequency, rate or extent of lyase activity, the catalysis of the cleavage of C-C, C-O, C-N and other bonds by other means than by hydrolysis or oxidation, or conversely adding a group to a double bond. They differ from other enzymes in that two substrates are involved in one reaction direction, but only one in the other direction. When acting on the single substrate, a molecule is eliminated and this generates either a new double bond or a new ring.
nucleobase, nucleoside and nucleotide metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides and nucleotides.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of molecular functions. Molecular functions are elemental biological activities occurring at the molecular level, such as catalysis or binding.
all
This term is the most general term possible
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cell proliferation
Any process that modulates the frequency, rate or extent of cell proliferation.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of adenylate cyclase activity
Any process that modulates the frequency, rate or extent of adenylate cyclase activity.
regulation of nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of nucleotides.
regulation of cyclase activity
Any process that modulates the frequency, rate or extent of cyclase activity.
regulation of cyclic nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of cyclic nucleotides.
regulation of nucleotide metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleotides.
regulation of adenylate cyclase activity
Any process that modulates the frequency, rate or extent of adenylate cyclase activity.
regulation of cAMP biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
regulation of nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of nucleotides.
cyclic nucleotide biosynthetic process
The chemical reactions and pathways resulting in the formation of a cyclic nucleotide, a nucleotide in which the phosphate group is in diester linkage to two positions on the sugar residue.
regulation of cyclic nucleotide metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving cyclic nucleotides.
regulation of cyclic nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of cyclic nucleotides.
cAMP biosynthetic process
The chemical reactions and pathways resulting in the formation of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
regulation of cAMP metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
regulation of cAMP biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04080 | 1.395e-02 | 0.8705 | 6 | 78 | Neuroactive ligand-receptor interaction |
ABHD5abhydrolase domain containing 5 (218739_at), score: -0.62 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.63 ADAM19ADAM metallopeptidase domain 19 (meltrin beta) (209765_at), score: -0.65 ADORA2Badenosine A2b receptor (205891_at), score: 0.86 AHI1Abelson helper integration site 1 (221569_at), score: -0.74 AIM1absent in melanoma 1 (212543_at), score: 0.84 AJAP1adherens junctions associated protein 1 (206460_at), score: -0.58 ANXA10annexin A10 (210143_at), score: -0.63 BAT1HLA-B associated transcript 1 (200041_s_at), score: 0.79 BMP6bone morphogenetic protein 6 (206176_at), score: -1 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.68 C2CD2C2 calcium-dependent domain containing 2 (212875_s_at), score: -0.72 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: -0.58 C5orf23chromosome 5 open reading frame 23 (219054_at), score: -0.68 CALB2calbindin 2 (205428_s_at), score: -0.85 CBLBCas-Br-M (murine) ecotropic retroviral transforming sequence b (209682_at), score: -0.59 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.72 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.8 CDCP1CUB domain containing protein 1 (218451_at), score: -0.6 CST6cystatin E/M (206595_at), score: -0.68 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.73 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.61 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.75 DIS3DIS3 mitotic control homolog (S. cerevisiae) (218362_s_at), score: -0.59 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.86 DUSP14dual specificity phosphatase 14 (203367_at), score: -0.67 DUSP4dual specificity phosphatase 4 (204014_at), score: -0.57 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: -0.63 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.82 ESRRAestrogen-related receptor alpha (1487_at), score: -0.61 FAM155Afamily with sequence similarity 155, member A (214825_at), score: -0.59 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: -0.71 FGF7fibroblast growth factor 7 (keratinocyte growth factor) (205782_at), score: -0.75 FICDFIC domain containing (219910_at), score: -0.62 FSTL3follistatin-like 3 (secreted glycoprotein) (203592_s_at), score: -0.59 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: -0.6 GKglycerol kinase (207387_s_at), score: -0.8 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.86 GRB14growth factor receptor-bound protein 14 (206204_at), score: -0.63 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.79 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: 0.81 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (207135_at), score: -0.82 IL11interleukin 11 (206924_at), score: -0.84 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.69 IL1RL1interleukin 1 receptor-like 1 (207526_s_at), score: -0.63 IL33interleukin 33 (209821_at), score: -0.81 INHBAinhibin, beta A (210511_s_at), score: -0.62 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.74 KCNG1potassium voltage-gated channel, subfamily G, member 1 (214595_at), score: -0.62 KLHL21kelch-like 21 (Drosophila) (203068_at), score: -0.66 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.65 MBOAT2membrane bound O-acyltransferase domain containing 2 (213288_at), score: -0.65 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.64 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.8 MREGmelanoregulin (219648_at), score: -0.64 NCALDneurocalcin delta (211685_s_at), score: -0.86 NEFMneurofilament, medium polypeptide (205113_at), score: -0.64 NOX4NADPH oxidase 4 (219773_at), score: -0.73 NPnucleoside phosphorylase (201695_s_at), score: -0.59 NTMneurotrimin (222020_s_at), score: -0.59 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.69 P2RX5purinergic receptor P2X, ligand-gated ion channel, 5 (210448_s_at), score: -0.67 PCDH9protocadherin 9 (219737_s_at), score: -0.81 PLAURplasminogen activator, urokinase receptor (211924_s_at), score: -0.59 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.62 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: -0.77 PTHLHparathyroid hormone-like hormone (211756_at), score: -0.9 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: -0.77 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.92 RNF44ring finger protein 44 (203286_at), score: 0.83 S1PR1sphingosine-1-phosphate receptor 1 (204642_at), score: -0.59 SCG5secretogranin V (7B2 protein) (203889_at), score: -0.62 SLC25A32solute carrier family 25, member 32 (221020_s_at), score: -0.59 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: -0.69 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.66 SMAD3SMAD family member 3 (218284_at), score: 0.91 SPA17sperm autoantigenic protein 17 (205406_s_at), score: 0.78 SSTR1somatostatin receptor 1 (208482_at), score: -0.58 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.83 TGFAtransforming growth factor, alpha (205016_at), score: -0.8 THYN1thymocyte nuclear protein 1 (218491_s_at), score: 0.78 TMEM2transmembrane protein 2 (218113_at), score: -0.76 TP53tumor protein p53 (201746_at), score: 0.8 TSPAN13tetraspanin 13 (217979_at), score: -0.67 XYLT1xylosyltransferase I (213725_x_at), score: -0.65
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690256.cel | 6 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949704.cel | 4 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |