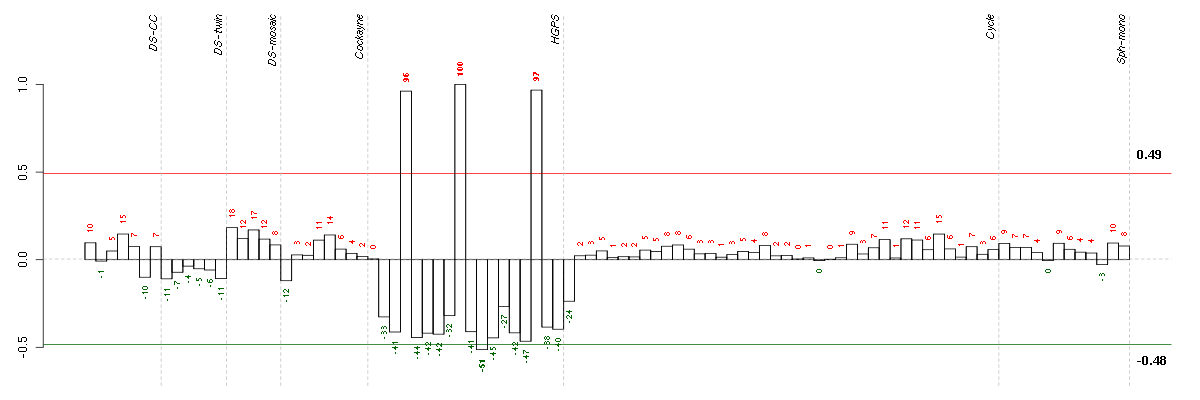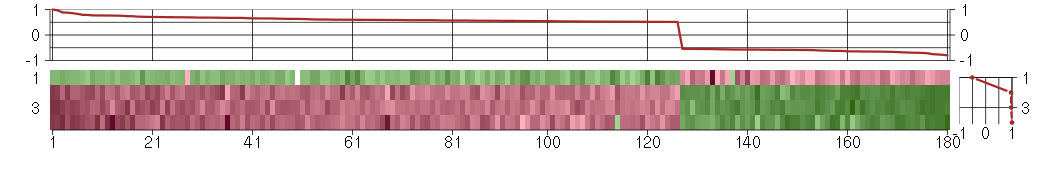

Under-expression is coded with green,
over-expression with red color.

cell motion
Any process involved in the controlled movement of a cell.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
locomotion
Self-propelled movement of a cell or organism from one location to another.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
localization of cell
Any process by which a cell is transported to, and/or maintained in, a specific location.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
cell motion
Any process involved in the controlled movement of a cell.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

calcium ion binding
Interacting selectively with calcium ions (Ca2+).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
structural molecule activity
The action of a molecule that contributes to the structural integrity of a complex or assembly within or outside a cell.
extracellular matrix structural constituent
The action of a molecule that contributes to the structural integrity of the extracellular matrix.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
metal ion binding
Interacting selectively with any metal ion.
ion binding
Interacting selectively with ions, charged atoms or groups of atoms.
cation binding
Interacting selectively with cations, charged atoms or groups of atoms with a net positive charge.
all
This term is the most general term possible
calcium ion binding
Interacting selectively with calcium ions (Ca2+).
ABCG2ATP-binding cassette, sub-family G (WHITE), member 2 (209735_at), score: 0.77 ACANaggrecan (205679_x_at), score: 0.65 ACTG2actin, gamma 2, smooth muscle, enteric (202274_at), score: 0.58 ADAM23ADAM metallopeptidase domain 23 (206046_at), score: 0.61 ADD3adducin 3 (gamma) (205882_x_at), score: 0.53 AGTR1angiotensin II receptor, type 1 (205357_s_at), score: 0.56 ANK3ankyrin 3, node of Ranvier (ankyrin G) (206385_s_at), score: 0.74 ANKRD6ankyrin repeat domain 6 (204671_s_at), score: 0.68 ANO3anoctamin 3 (215241_at), score: 0.51 APOL3apolipoprotein L, 3 (221087_s_at), score: 0.53 APOLD1apolipoprotein L domain containing 1 (221031_s_at), score: 0.59 AQP1aquaporin 1 (Colton blood group) (209047_at), score: 0.67 ARHGAP29Rho GTPase activating protein 29 (203910_at), score: -0.67 ARHGDIBRho GDP dissociation inhibitor (GDI) beta (201288_at), score: 0.57 ARHGEF10LRho guanine nucleotide exchange factor (GEF) 10-like (221656_s_at), score: 0.53 ARL4CADP-ribosylation factor-like 4C (202207_at), score: 0.68 ATF3activating transcription factor 3 (202672_s_at), score: 0.7 BNC2basonuclin 2 (220272_at), score: -0.57 BTN3A2butyrophilin, subfamily 3, member A2 (209846_s_at), score: 0.58 BTN3A3butyrophilin, subfamily 3, member A3 (204820_s_at), score: 0.61 C16orf45chromosome 16 open reading frame 45 (212736_at), score: 0.55 C21orf7chromosome 21 open reading frame 7 (221211_s_at), score: 0.52 C3complement component 3 (217767_at), score: -0.55 CAPGcapping protein (actin filament), gelsolin-like (201850_at), score: -0.56 CARHSP1calcium regulated heat stable protein 1, 24kDa (218384_at), score: -0.62 CCDC81coiled-coil domain containing 81 (220389_at), score: 0.7 CCDC92coiled-coil domain containing 92 (218175_at), score: 0.51 CCND2cyclin D2 (200953_s_at), score: 0.56 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.69 CDK5R1cyclin-dependent kinase 5, regulatory subunit 1 (p35) (204995_at), score: 0.53 CLEC2BC-type lectin domain family 2, member B (209732_at), score: -0.59 CLEC3BC-type lectin domain family 3, member B (205200_at), score: 0.59 CLIC3chloride intracellular channel 3 (219529_at), score: -0.55 COL4A5collagen, type IV, alpha 5 (213110_s_at), score: 0.61 CORINcorin, serine peptidase (220356_at), score: 0.75 CRLF1cytokine receptor-like factor 1 (206315_at), score: -0.55 CXCL12chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) (209687_at), score: 0.52 CYB5Acytochrome b5 type A (microsomal) (215726_s_at), score: -0.66 CYFIP2cytoplasmic FMR1 interacting protein 2 (215785_s_at), score: 0.76 DAAM2dishevelled associated activator of morphogenesis 2 (212793_at), score: 0.71 DPYSL3dihydropyrimidinase-like 3 (201431_s_at), score: -0.58 EFHD1EF-hand domain family, member D1 (209343_at), score: 0.76 EHD3EH-domain containing 3 (218935_at), score: 0.6 EMP1epithelial membrane protein 1 (201325_s_at), score: -0.62 ENDOD1endonuclease domain containing 1 (212573_at), score: 0.55 ENPEPglutamyl aminopeptidase (aminopeptidase A) (204844_at), score: 0.65 EREGepiregulin (205767_at), score: 0.59 FBLN1fibulin 1 (202995_s_at), score: -0.65 FBLN2fibulin 2 (203886_s_at), score: 0.52 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.56 FLT3LGfms-related tyrosine kinase 3 ligand (210607_at), score: 0.52 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: 0.69 FRMD4AFERM domain containing 4A (208476_s_at), score: 0.52 FUT8fucosyltransferase 8 (alpha (1,6) fucosyltransferase) (203988_s_at), score: -0.68 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: 0.58 GALNT3UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) (203397_s_at), score: 0.71 GPR116G protein-coupled receptor 116 (212950_at), score: 1 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: 0.65 GREM2gremlin 2, cysteine knot superfamily, homolog (Xenopus laevis) (220794_at), score: -0.59 GRIK2glutamate receptor, ionotropic, kainate 2 (213845_at), score: 0.51 GTDC1glycosyltransferase-like domain containing 1 (219770_at), score: 0.55 HCLS1hematopoietic cell-specific Lyn substrate 1 (202957_at), score: 0.56 HNMThistamine N-methyltransferase (204112_s_at), score: 0.66 HOXB7homeobox B7 (204779_s_at), score: -0.7 HSPB2heat shock 27kDa protein 2 (205824_at), score: 0.56 HSPB8heat shock 22kDa protein 8 (221667_s_at), score: 0.55 ID1inhibitor of DNA binding 1, dominant negative helix-loop-helix protein (208937_s_at), score: -0.72 IRF1interferon regulatory factor 1 (202531_at), score: 0.6 IRS2insulin receptor substrate 2 (209185_s_at), score: -0.57 ITGA6integrin, alpha 6 (215177_s_at), score: -0.57 ITGA8integrin, alpha 8 (214265_at), score: 0.89 IVNS1ABPinfluenza virus NS1A binding protein (206245_s_at), score: -0.59 KALRNkalirin, RhoGEF kinase (206078_at), score: 0.7 KCNE4potassium voltage-gated channel, Isk-related family, member 4 (222379_at), score: 0.67 KCTD12potassium channel tetramerisation domain containing 12 (212192_at), score: -0.63 KIAA0355KIAA0355 (203288_at), score: 0.57 KIRRELkin of IRRE like (Drosophila) (220825_s_at), score: 0.53 KITv-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (205051_s_at), score: 0.52 KLF5Kruppel-like factor 5 (intestinal) (209212_s_at), score: -0.66 LAMA4laminin, alpha 4 (202202_s_at), score: -0.8 LGR5leucine-rich repeat-containing G protein-coupled receptor 5 (213880_at), score: 0.7 LIMCH1LIM and calponin homology domains 1 (212327_at), score: -0.65 LMO4LIM domain only 4 (209205_s_at), score: -0.65 LMOD1leiomodin 1 (smooth muscle) (203766_s_at), score: 0.64 LPPR4plasticity related gene 1 (213496_at), score: 0.68 LRRC32leucine rich repeat containing 32 (203835_at), score: 0.63 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: -0.6 MAPRE2microtubule-associated protein, RP/EB family, member 2 (202501_at), score: 0.58 MATN3matrilin 3 (206091_at), score: -0.58 MBPmyelin basic protein (210136_at), score: 0.63 MMDmonocyte to macrophage differentiation-associated (203414_at), score: -0.58 MMP2matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (201069_at), score: -0.77 MRASmuscle RAS oncogene homolog (206538_at), score: -0.57 MYBL1v-myb myeloblastosis viral oncogene homolog (avian)-like 1 (213906_at), score: -0.59 MYH10myosin, heavy chain 10, non-muscle (212372_at), score: -0.76 MYO1Emyosin IE (203072_at), score: 0.57 NAAAN-acylethanolamine acid amidase (214765_s_at), score: 0.52 NAGLUN-acetylglucosaminidase, alpha- (204360_s_at), score: 0.54 NAP1L3nucleosome assembly protein 1-like 3 (204749_at), score: -0.65 NBEAneurobeachin (221207_s_at), score: 0.58 NEFMneurofilament, medium polypeptide (205113_at), score: 0.77 NESnestin (218678_at), score: 0.58 NLRP1NLR family, pyrin domain containing 1 (218380_at), score: 0.52 NMUneuromedin U (206023_at), score: 0.54 NOTCH1Notch homolog 1, translocation-associated (Drosophila) (218902_at), score: 0.54 NRP2neuropilin 2 (214632_at), score: -0.58 NRXN3neurexin 3 (205795_at), score: 0.69 PARP3poly (ADP-ribose) polymerase family, member 3 (209940_at), score: 0.6 PARP8poly (ADP-ribose) polymerase family, member 8 (219033_at), score: 0.51 PDGFAplatelet-derived growth factor alpha polypeptide (205463_s_at), score: 0.65 PDLIM3PDZ and LIM domain 3 (209621_s_at), score: 0.74 PELOpelota homolog (Drosophila) (218472_s_at), score: 0.52 PHF11PHD finger protein 11 (221816_s_at), score: 0.57 PIK3R1phosphoinositide-3-kinase, regulatory subunit 1 (alpha) (212240_s_at), score: 0.53 PKIAprotein kinase (cAMP-dependent, catalytic) inhibitor alpha (204612_at), score: -0.57 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.6 PLD1phospholipase D1, phosphatidylcholine-specific (215723_s_at), score: 0.6 PLK2polo-like kinase 2 (Drosophila) (201939_at), score: -0.7 PLXDC1plexin domain containing 1 (219700_at), score: 0.57 PORCNporcupine homolog (Drosophila) (219483_s_at), score: 0.79 PPP1R13Lprotein phosphatase 1, regulatory (inhibitor) subunit 13 like (218849_s_at), score: -0.65 PPT2palmitoyl-protein thioesterase 2 (209490_s_at), score: 0.52 PRKAG2protein kinase, AMP-activated, gamma 2 non-catalytic subunit (218292_s_at), score: 0.54 PROCRprotein C receptor, endothelial (EPCR) (203650_at), score: 0.72 PRPS1phosphoribosyl pyrophosphate synthetase 1 (209440_at), score: -0.59 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: 0.68 PRSS3protease, serine, 3 (213421_x_at), score: 0.79 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: 0.57 PTK2PTK2 protein tyrosine kinase 2 (207821_s_at), score: 0.52 PTPRZ1protein tyrosine phosphatase, receptor-type, Z polypeptide 1 (204469_at), score: 0.97 RELNreelin (205923_at), score: 0.56 RHBDL2rhomboid, veinlet-like 2 (Drosophila) (219489_s_at), score: -0.75 RPP25ribonuclease P/MRP 25kDa subunit (219143_s_at), score: 0.72 RTN1reticulon 1 (203485_at), score: 0.88 S100A3S100 calcium binding protein A3 (206027_at), score: 0.6 SCN3Asodium channel, voltage-gated, type III, alpha subunit (210432_s_at), score: 0.67 SEPT6septin 6 (212414_s_at), score: -0.61 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: -0.61 SERPINF1serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 (202283_at), score: -0.57 SIGLEC15sialic acid binding Ig-like lectin 15 (215856_at), score: 0.71 SIM2single-minded homolog 2 (Drosophila) (206558_at), score: 0.86 SIX2SIX homeobox 2 (206510_at), score: 0.68 SLC22A17solute carrier family 22, member 17 (218675_at), score: -0.64 SLC24A3solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 (57588_at), score: 0.83 SLC7A5solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 (201195_s_at), score: -0.64 SMAD6SMAD family member 6 (207069_s_at), score: -0.58 SNTB1syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) (208608_s_at), score: 0.59 SORDsorbitol dehydrogenase (201563_at), score: -0.56 SORT1sortilin 1 (212807_s_at), score: 0.55 SPINT2serine peptidase inhibitor, Kunitz type, 2 (210715_s_at), score: 0.58 SPP1secreted phosphoprotein 1 (209875_s_at), score: 0.67 STAT4signal transducer and activator of transcription 4 (206118_at), score: 0.63 STYK1serine/threonine/tyrosine kinase 1 (221696_s_at), score: 0.53 SYNMsynemin, intermediate filament protein (212730_at), score: 0.65 TBC1D4TBC1 domain family, member 4 (203386_at), score: -0.55 TBXA2Rthromboxane A2 receptor (336_at), score: 0.7 TEAD3TEA domain family member 3 (209454_s_at), score: 0.62 TFPItissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) (209676_at), score: 0.58 TINAGL1tubulointerstitial nephritis antigen-like 1 (219058_x_at), score: 0.63 TMEM35transmembrane protein 35 (219685_at), score: 0.56 TNFSF4tumor necrosis factor (ligand) superfamily, member 4 (207426_s_at), score: 0.59 TNXAtenascin XA pseudogene (213451_x_at), score: 0.55 TNXBtenascin XB (216333_x_at), score: 0.59 TRIM2tripartite motif-containing 2 (202342_s_at), score: -0.64 TRIM34tripartite motif-containing 34 (221044_s_at), score: 0.67 TRPV2transient receptor potential cation channel, subfamily V, member 2 (219282_s_at), score: 0.64 TRY6trypsinogen C (215395_x_at), score: 0.76 TSPAN12tetraspanin 12 (219274_at), score: 0.59 TSPYL5TSPY-like 5 (213122_at), score: -0.69 UAP1UDP-N-acteylglucosamine pyrophosphorylase 1 (209340_at), score: -0.54 UAP1L1UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 (214755_at), score: 0.55 UBA7ubiquitin-like modifier activating enzyme 7 (203281_s_at), score: 0.61 VAMP8vesicle-associated membrane protein 8 (endobrevin) (202546_at), score: 0.77 WWP2WW domain containing E3 ubiquitin protein ligase 2 (204022_at), score: 0.53 ZEB2zinc finger E-box binding homeobox 2 (203603_s_at), score: 0.6 ZFHX4zinc finger homeobox 4 (219779_at), score: -0.58 ZFP36zinc finger protein 36, C3H type, homolog (mouse) (201531_at), score: 0.52 ZFPM2zinc finger protein, multitype 2 (219778_at), score: -0.55 ZNF185zinc finger protein 185 (LIM domain) (203585_at), score: -0.55
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690352.cel | 11 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690336.cel | 9 | 5 | HGPS | hgu133a | HGPS | AG10750 |