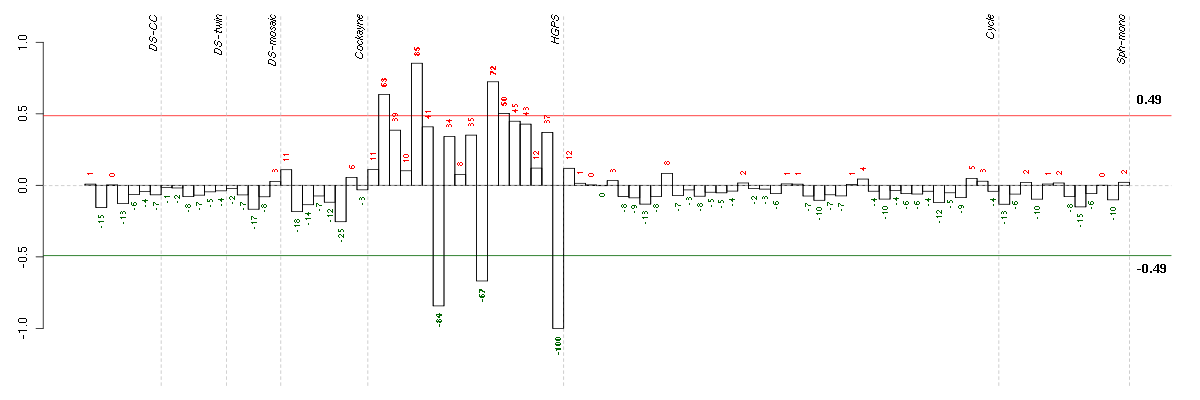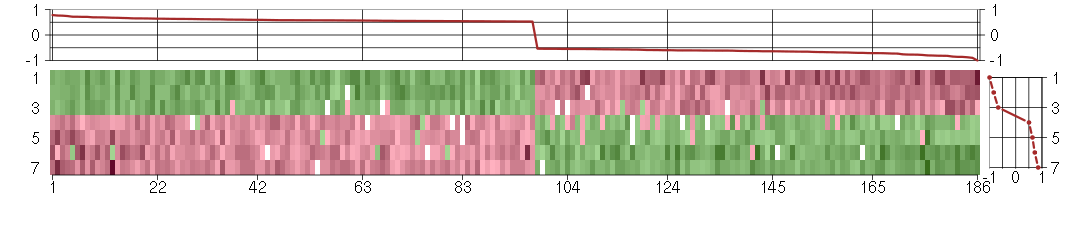

Under-expression is coded with green,
over-expression with red color.

defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
muscle development
The process whose specific outcome is the progression of the muscle over time, from its formation to the mature structure. The muscle is an organ consisting of a tissue made up of various elongated cells that are specialized to contract and thus to produce movement and mechanical work.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
proteinaceous extracellular matrix
A layer consisting mainly of proteins (especially collagen) and glycosaminoglycans (mostly as proteoglycans) that forms a sheet underlying or overlying cells such as endothelial and epithelial cells. The proteins are secreted by cells in the vicinity.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

calcium ion binding
Interacting selectively with calcium ions (Ca2+).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
carbohydrate binding
Interacting selectively with any carbohydrate.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
metal ion binding
Interacting selectively with any metal ion.
ion binding
Interacting selectively with ions, charged atoms or groups of atoms.
cation binding
Interacting selectively with cations, charged atoms or groups of atoms with a net positive charge.
all
This term is the most general term possible
calcium ion binding
Interacting selectively with calcium ions (Ca2+).
ABCA8ATP-binding cassette, sub-family A (ABC1), member 8 (204719_at), score: -0.68 ACTC1actin, alpha, cardiac muscle 1 (205132_at), score: 0.57 ADAMTS3ADAM metallopeptidase with thrombospondin type 1 motif, 3 (214913_at), score: -0.61 ADAMTSL3ADAMTS-like 3 (213974_at), score: -0.77 ADMadrenomedullin (202912_at), score: 0.54 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: -0.7 AMFRautocrine motility factor receptor (202203_s_at), score: -0.63 ANGPT1angiopoietin 1 (205608_s_at), score: 0.55 ANKRD1ankyrin repeat domain 1 (cardiac muscle) (206029_at), score: -0.78 ANTXR1anthrax toxin receptor 1 (220092_s_at), score: 0.57 ANXA10annexin A10 (210143_at), score: -0.61 ANXA3annexin A3 (209369_at), score: -0.55 ARHGEF17Rho guanine nucleotide exchange factor (GEF) 17 (203756_at), score: 0.55 ARSBarylsulfatase B (206129_s_at), score: -0.68 B3GALT4UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 (210205_at), score: 0.54 BNC2basonuclin 2 (220272_at), score: -0.6 C11orf63chromosome 11 open reading frame 63 (220141_at), score: 0.64 C16orf5chromosome 16 open reading frame 5 (218183_at), score: 0.57 C1orf54chromosome 1 open reading frame 54 (219506_at), score: -0.56 C4orf31chromosome 4 open reading frame 31 (219747_at), score: 0.67 C8orf84chromosome 8 open reading frame 84 (214725_at), score: -0.87 CADPS2Ca++-dependent secretion activator 2 (219572_at), score: -0.61 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.55 CCL2chemokine (C-C motif) ligand 2 (216598_s_at), score: -0.55 CD200CD200 molecule (209583_s_at), score: -0.77 CD248CD248 molecule, endosialin (219025_at), score: 0.55 CD34CD34 molecule (209543_s_at), score: -0.81 CD9CD9 molecule (201005_at), score: -0.54 CEBPGCCAAT/enhancer binding protein (C/EBP), gamma (204203_at), score: -0.54 CFHcomplement factor H (213800_at), score: -0.68 CFHR1complement factor H-related 1 (215388_s_at), score: -0.55 CGBchorionic gonadotropin, beta polypeptide (205387_s_at), score: 0.71 CHN1chimerin (chimaerin) 1 (212624_s_at), score: 0.74 CHRDL1chordin-like 1 (209763_at), score: -0.59 CLDN1claudin 1 (218182_s_at), score: -0.58 COMPcartilage oligomeric matrix protein (205713_s_at), score: 0.56 CRYBB2crystallin, beta B2 (206777_s_at), score: -0.63 CSDC2cold shock domain containing C2, RNA binding (209981_at), score: 0.54 CXCL1chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) (204470_at), score: -0.71 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.59 CYP26B1cytochrome P450, family 26, subfamily B, polypeptide 1 (219825_at), score: 0.69 DAAM1dishevelled associated activator of morphogenesis 1 (216060_s_at), score: -0.65 DAB2disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) (201279_s_at), score: 0.59 DBN1drebrin 1 (217025_s_at), score: 0.59 DDX43DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 (220004_at), score: -0.64 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.56 DSG2desmoglein 2 (217901_at), score: -0.64 DSTdystonin (212253_x_at), score: 0.72 DUSP4dual specificity phosphatase 4 (204014_at), score: -0.57 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.58 EDN1endothelin 1 (218995_s_at), score: -0.55 ELL2elongation factor, RNA polymerase II, 2 (214446_at), score: 0.53 ELTD1EGF, latrophilin and seven transmembrane domain containing 1 (219134_at), score: 0.56 EN1engrailed homeobox 1 (220559_at), score: 0.55 EPHA4EPH receptor A4 (206114_at), score: 0.57 EPHA5EPH receptor A5 (215664_s_at), score: -0.86 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (207347_at), score: -0.62 ERGv-ets erythroblastosis virus E26 oncogene homolog (avian) (213541_s_at), score: -0.85 F3coagulation factor III (thromboplastin, tissue factor) (204363_at), score: 0.54 F8coagulation factor VIII, procoagulant component (205756_s_at), score: 0.53 FAM65Bfamily with sequence similarity 65, member B (209829_at), score: 0.62 FGL2fibrinogen-like 2 (204834_at), score: -1 FMO3flavin containing monooxygenase 3 (40665_at), score: -0.71 FOXL2forkhead box L2 (220102_at), score: 0.53 GABRA5gamma-aminobutyric acid (GABA) A receptor, alpha 5 (206456_at), score: 0.78 GALNT1UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) (201723_s_at), score: -0.56 GALNT12UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) (218885_s_at), score: -0.6 GALNT7UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) (218313_s_at), score: -0.55 GATA6GATA binding protein 6 (210002_at), score: -0.61 GPR126G protein-coupled receptor 126 (213094_at), score: -0.61 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: -0.65 GRAMD3GRAM domain containing 3 (218706_s_at), score: 0.65 GRPgastrin-releasing peptide (206326_at), score: 0.63 GSTT2glutathione S-transferase theta 2 (205439_at), score: -0.71 HABP4hyaluronan binding protein 4 (209818_s_at), score: 0.62 HDAC5histone deacetylase 5 (202455_at), score: 0.6 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.55 HOXB2homeobox B2 (205453_at), score: 0.54 HOXB6homeobox B6 (205366_s_at), score: 0.64 HSPB8heat shock 22kDa protein 8 (221667_s_at), score: 0.58 HYIhydroxypyruvate isomerase homolog (E. coli) (221435_x_at), score: -0.76 IFI27interferon, alpha-inducible protein 27 (202411_at), score: -0.61 IL18interleukin 18 (interferon-gamma-inducing factor) (206295_at), score: -0.69 IL1Ainterleukin 1, alpha (210118_s_at), score: -0.64 INF2inverted formin, FH2 and WH2 domain containing (218144_s_at), score: 0.53 ITM2Aintegral membrane protein 2A (202746_at), score: -0.7 KIAA0182KIAA0182 (212057_at), score: 0.65 KRT34keratin 34 (206969_at), score: 0.55 KRTAP1-1keratin associated protein 1-1 (220976_s_at), score: 0.76 LAMC2laminin, gamma 2 (202267_at), score: -0.82 LARGElike-glycosyltransferase (215543_s_at), score: 0.56 LBHlimb bud and heart development homolog (mouse) (221011_s_at), score: 0.69 LGALS3BPlectin, galactoside-binding, soluble, 3 binding protein (200923_at), score: -0.56 LIPGlipase, endothelial (219181_at), score: -0.9 LMO2LIM domain only 2 (rhombotin-like 1) (204249_s_at), score: -0.6 LOC100132214similar to HSPC047 protein (220692_at), score: -0.6 LYPD1LY6/PLAUR domain containing 1 (212909_at), score: -0.8 MAPK3mitogen-activated protein kinase 3 (212046_x_at), score: 0.55 MCTP1multiple C2 domains, transmembrane 1 (220122_at), score: -0.69 MEF2Cmyocyte enhancer factor 2C (209199_s_at), score: 0.55 MEGF6multiple EGF-like-domains 6 (213942_at), score: 0.63 MFAP2microfibrillar-associated protein 2 (203417_at), score: 0.59 MFSD10major facilitator superfamily domain containing 10 (209215_at), score: 0.58 MGPmatrix Gla protein (202291_s_at), score: -0.71 MMP1matrix metallopeptidase 1 (interstitial collagenase) (204475_at), score: -0.8 MOSPD3motile sperm domain containing 3 (219070_s_at), score: 0.57 MT1Mmetallothionein 1M (217546_at), score: 0.6 MVPmajor vault protein (202180_s_at), score: 0.58 MYCT1myc target 1 (220471_s_at), score: -0.64 MYLKmyosin light chain kinase (202555_s_at), score: 0.71 NCAM1neural cell adhesion molecule 1 (212843_at), score: 0.53 NFATC3nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 (210555_s_at), score: -0.57 NFIBnuclear factor I/B (209290_s_at), score: -0.66 NOX4NADPH oxidase 4 (219773_at), score: -0.73 NRG1neuregulin 1 (206343_s_at), score: 0.53 OXTRoxytocin receptor (206825_at), score: 0.63 PARP3poly (ADP-ribose) polymerase family, member 3 (209940_at), score: 0.54 PDE10Aphosphodiesterase 10A (205501_at), score: -0.65 PDE4Bphosphodiesterase 4B, cAMP-specific (phosphodiesterase E4 dunce homolog, Drosophila) (203708_at), score: 0.55 PDZRN4PDZ domain containing ring finger 4 (220595_at), score: -0.65 PELI1pellino homolog 1 (Drosophila) (218319_at), score: -0.54 PKP2plakophilin 2 (207717_s_at), score: -0.54 PLA2G4Aphospholipase A2, group IVA (cytosolic, calcium-dependent) (210145_at), score: -0.63 PLAC8placenta-specific 8 (219014_at), score: 0.65 PLCB4phospholipase C, beta 4 (203896_s_at), score: 0.62 PLCL1phospholipase C-like 1 (205934_at), score: -0.68 PLXNC1plexin C1 (213241_at), score: 0.65 POPDC3popeye domain containing 3 (219926_at), score: 0.67 PPAP2Aphosphatidic acid phosphatase type 2A (209147_s_at), score: 0.61 PPM1Hprotein phosphatase 1H (PP2C domain containing) (212686_at), score: -0.77 PRKD1protein kinase D1 (205880_at), score: 0.6 PSG5pregnancy specific beta-1-glycoprotein 5 (204830_x_at), score: 0.6 PSG6pregnancy specific beta-1-glycoprotein 6 (209738_x_at), score: 0.6 PSG9pregnancy specific beta-1-glycoprotein 9 (209594_x_at), score: 0.57 PSPHphosphoserine phosphatase (205048_s_at), score: -0.6 PTGER3prostaglandin E receptor 3 (subtype EP3) (213933_at), score: 0.61 PTPLBprotein tyrosine phosphatase-like (proline instead of catalytic arginine), member b (212640_at), score: 0.56 RAC2ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) (213603_s_at), score: 0.6 RARBretinoic acid receptor, beta (205080_at), score: 0.72 RGNregucalcin (senescence marker protein-30) (210751_s_at), score: 0.58 RGS7regulator of G-protein signaling 7 (206290_s_at), score: -0.6 ROR1receptor tyrosine kinase-like orphan receptor 1 (205805_s_at), score: -0.63 RPH3ALrabphilin 3A-like (without C2 domains) (221614_s_at), score: 0.58 RUNX3runt-related transcription factor 3 (204198_s_at), score: 0.66 SALL1sal-like 1 (Drosophila) (206893_at), score: 0.68 SCRN1secernin 1 (201462_at), score: 0.53 SDC3syndecan 3 (202898_at), score: 0.58 SERPING1serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 (200986_at), score: 0.54 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: 0.53 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: 0.69 SLC17A9solute carrier family 17, member 9 (219559_at), score: 0.56 SLC25A1solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 (210010_s_at), score: 0.53 SLC31A2solute carrier family 31 (copper transporters), member 2 (204204_at), score: -0.61 SLC39A8solute carrier family 39 (zinc transporter), member 8 (209267_s_at), score: -0.73 SLC7A8solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 (216092_s_at), score: 0.58 SLC9A3R2solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 (209830_s_at), score: 0.63 SNTA1syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component) (203516_at), score: 0.57 SOCS2suppressor of cytokine signaling 2 (203373_at), score: 0.6 SOX17SRY (sex determining region Y)-box 17 (219993_at), score: -0.86 STBD1starch binding domain 1 (203986_at), score: 0.55 STMN2stathmin-like 2 (203000_at), score: 0.76 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.61 SUSD5sushi domain containing 5 (214954_at), score: -0.57 SVILsupervillin (202565_s_at), score: -0.54 TADA3Ltranscriptional adaptor 3 (NGG1 homolog, yeast)-like (215273_s_at), score: 0.63 TBC1D17TBC1 domain family, member 17 (218466_at), score: 0.55 THY1Thy-1 cell surface antigen (208850_s_at), score: 0.64 TIMP3TIMP metallopeptidase inhibitor 3 (201149_s_at), score: 0.57 TJP2tight junction protein 2 (zona occludens 2) (202085_at), score: -0.57 TLR4toll-like receptor 4 (221060_s_at), score: -0.81 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: -0.65 TNFAIP8tumor necrosis factor, alpha-induced protein 8 (210260_s_at), score: -0.72 TNFRSF6Btumor necrosis factor receptor superfamily, member 6b, decoy (206467_x_at), score: 0.68 TOXthymocyte selection-associated high mobility group box (204529_s_at), score: -0.56 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: -0.67 USP5ubiquitin specific peptidase 5 (isopeptidase T) (206031_s_at), score: 0.54 VLDLRvery low density lipoprotein receptor (209822_s_at), score: -0.56 VPS13Avacuolar protein sorting 13 homolog A (S. cerevisiae) (214785_at), score: -0.64 WNT5Bwingless-type MMTV integration site family, member 5B (221029_s_at), score: 0.58 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.63 ZFHX4zinc finger homeobox 4 (219779_at), score: -0.59 ZMIZ1zinc finger, MIZ-type containing 1 (212124_at), score: 0.58 ZNF323zinc finger protein 323 (222016_s_at), score: -0.67 ZNF365zinc finger protein 365 (206448_at), score: -0.55 ZNF423zinc finger protein 423 (214761_at), score: -0.61 ZNF711zinc finger protein 711 (207781_s_at), score: 0.62
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690480.cel | 18 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690352.cel | 11 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690376.cel | 13 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-3860-raw-cel-1561690215.cel | 2 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |