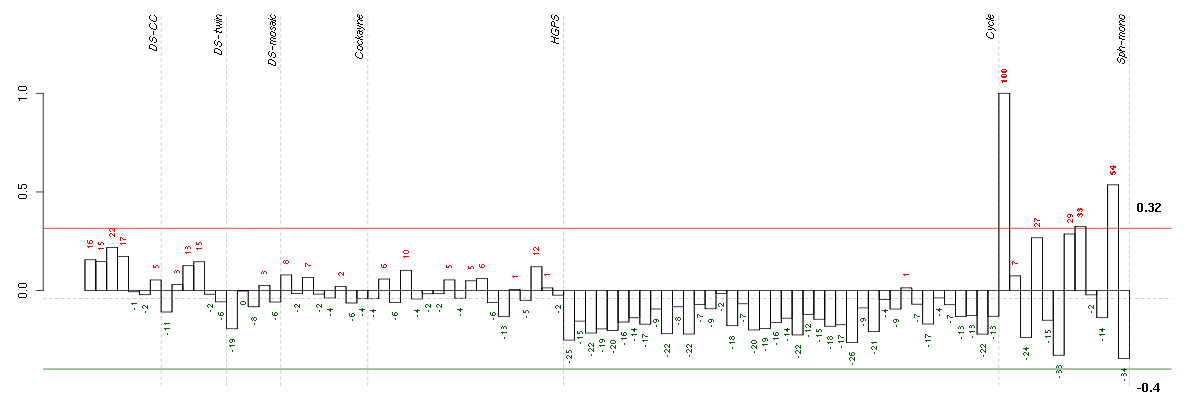

Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
nucleic acid binding
Interacting selectively with any nucleic acid.
DNA binding
Interacting selectively with DNA (deoxyribonucleic acid).
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
all
This term is the most general term possible
ABCA1ATP-binding cassette, sub-family A (ABC1), member 1 (203504_s_at), score: 0.6 ABCG1ATP-binding cassette, sub-family G (WHITE), member 1 (204567_s_at), score: 0.79 ACOT11acyl-CoA thioesterase 11 (216103_at), score: 0.64 ADORA2Aadenosine A2a receptor (205013_s_at), score: 1 AKAP8A kinase (PRKA) anchor protein 8 (203848_at), score: 0.64 ALDH1B1aldehyde dehydrogenase 1 family, member B1 (209646_x_at), score: 0.61 ALDOAP2aldolase A, fructose-bisphosphate pseudogene 2 (211617_at), score: 0.63 ALMS1Alstrom syndrome 1 (214707_x_at), score: 0.87 AMPD3adenosine monophosphate deaminase (isoform E) (207992_s_at), score: 0.6 ANKHankylosis, progressive homolog (mouse) (220076_at), score: 0.7 ANKRD10ankyrin repeat domain 10 (218093_s_at), score: 0.75 APBB2amyloid beta (A4) precursor protein-binding, family B, member 2 (212972_x_at), score: 0.92 APODapolipoprotein D (201525_at), score: 0.7 ARAP2ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 (214102_at), score: 0.67 ARHGEF7Rho guanine nucleotide exchange factor (GEF) 7 (202548_s_at), score: 0.67 ARID4AAT rich interactive domain 4A (RBP1-like) (205062_x_at), score: 0.62 BACH2BTB and CNC homology 1, basic leucine zipper transcription factor 2 (221234_s_at), score: 0.61 BCL9B-cell CLL/lymphoma 9 (204129_at), score: 0.77 BHLHB9basic helix-loop-helix domain containing, class B, 9 (213709_at), score: 0.86 BTAF1BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa (Mot1 homolog, S. cerevisiae) (209430_at), score: 0.63 C11orf57chromosome 11 open reading frame 57 (218314_s_at), score: 0.72 C13orf23chromosome 13 open reading frame 23 (218420_s_at), score: 0.7 C17orf86chromosome 17 open reading frame 86 (221621_at), score: 0.68 C4orf10chromosome 4 open reading frame 10 (214123_s_at), score: 0.74 C6orf106chromosome 6 open reading frame 106 (217925_s_at), score: 0.59 CBFA2T2core-binding factor, runt domain, alpha subunit 2; translocated to, 2 (207625_s_at), score: 0.61 CDC14ACDC14 cell division cycle 14 homolog A (S. cerevisiae) (205288_at), score: 0.8 CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213183_s_at), score: 0.76 CDR1cerebellar degeneration-related protein 1, 34kDa (207276_at), score: 0.74 CGGBP1CGG triplet repeat binding protein 1 (214050_at), score: 0.89 CHI3L1chitinase 3-like 1 (cartilage glycoprotein-39) (209395_at), score: 0.79 CIRBPcold inducible RNA binding protein (200811_at), score: 0.67 CISD3CDGSH iron sulfur domain 3 (213551_x_at), score: 0.66 COX5Bcytochrome c oxidase subunit Vb (213736_at), score: 0.61 CPEcarboxypeptidase E (201117_s_at), score: 0.79 CXCL1chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) (204470_at), score: 0.61 CXCL2chemokine (C-X-C motif) ligand 2 (209774_x_at), score: 0.81 CYP1A2cytochrome P450, family 1, subfamily A, polypeptide 2 (207608_x_at), score: 0.6 CYP2B6cytochrome P450, family 2, subfamily B, polypeptide 6 (217133_x_at), score: 0.6 DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (219290_x_at), score: 0.84 DAZ1deleted in azoospermia 1 (216922_x_at), score: 0.63 DAZ4deleted in azoospermia 4 (216351_x_at), score: 0.63 DIP2ADIP2 disco-interacting protein 2 homolog A (Drosophila) (215529_x_at), score: 0.63 DKFZp686O1327hypothetical gene supported by BC043549; BX648102 (216877_at), score: 0.79 DNAH3dynein, axonemal, heavy chain 3 (220725_x_at), score: 0.9 EEF1Deukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) (214395_x_at), score: 0.78 EIF5A2eukaryotic translation initiation factor 5A2 (220198_s_at), score: 0.62 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (206919_at), score: 0.71 EPB41L1erythrocyte membrane protein band 4.1-like 1 (212339_at), score: 0.6 ETS2v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) (201328_at), score: 0.63 FAM134Afamily with sequence similarity 134, member A (222129_at), score: 0.69 FAM60Afamily with sequence similarity 60, member A (220147_s_at), score: 0.59 FBXL11F-box and leucine-rich repeat protein 11 (208989_s_at), score: 0.7 FBXW12F-box and WD repeat domain containing 12 (215600_x_at), score: 0.78 FCARFc fragment of IgA, receptor for (211305_x_at), score: 0.73 FGD6FYVE, RhoGEF and PH domain containing 6 (219901_at), score: 0.71 FLJ10038hypothetical protein FLJ10038 (205510_s_at), score: 0.71 FLJ11292hypothetical protein FLJ11292 (220828_s_at), score: 0.91 FLJ23172hypothetical LOC389177 (217016_x_at), score: 0.7 FLJ42627hypothetical LOC645644 (220352_x_at), score: 0.6 FNBP4formin binding protein 4 (212232_at), score: 0.74 FOXC1forkhead box C1 (213260_at), score: 0.64 FUBP1far upstream element (FUSE) binding protein 1 (212847_at), score: 0.73 G3BP1GTPase activating protein (SH3 domain) binding protein 1 (222187_x_at), score: 0.92 GCLCglutamate-cysteine ligase, catalytic subunit (202922_at), score: 0.59 GORASP1golgi reassembly stacking protein 1, 65kDa (56919_at), score: 0.72 GPM6Bglycoprotein M6B (209167_at), score: 0.82 GTPBP3GTP binding protein 3 (mitochondrial) (213835_x_at), score: 0.68 H2AFJH2A histone family, member J (220936_s_at), score: 0.59 HCG2P7HLA complex group 2 pseudogene 7 (216229_x_at), score: 0.79 HECAheadcase homolog (Drosophila) (218603_at), score: 0.71 HEY1hairy/enhancer-of-split related with YRPW motif 1 (44783_s_at), score: 0.65 HIST2H2BEhistone cluster 2, H2be (202708_s_at), score: 0.77 HNRNPDheterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) (213359_at), score: 0.85 HS2ST1heparan sulfate 2-O-sulfotransferase 1 (203285_s_at), score: 0.63 IBD12Inflammatory bowel disease 12 (215373_x_at), score: 0.82 IER3IP1immediate early response 3 interacting protein 1 (211406_at), score: 0.66 IL1RAPinterleukin 1 receptor accessory protein (205227_at), score: 0.7 IL1RNinterleukin 1 receptor antagonist (212657_s_at), score: 0.64 IL24interleukin 24 (206569_at), score: 0.64 INSRinsulin receptor (213792_s_at), score: 0.86 IREB2iron-responsive element binding protein 2 (214666_x_at), score: 0.67 ISL1ISL LIM homeobox 1 (206104_at), score: 0.68 ITGB5integrin, beta 5 (214021_x_at), score: 0.68 ITPKBinositol 1,4,5-trisphosphate 3-kinase B (203723_at), score: 0.71 JARID2jumonji, AT rich interactive domain 2 (203297_s_at), score: 0.66 KIAA0894KIAA0894 protein (207436_x_at), score: 0.67 KLF11Kruppel-like factor 11 (218486_at), score: 0.65 LOC100128701heterogeneous nuclear ribonucleoprotein A1-like 2 pseudogene (216497_at), score: 0.82 LOC100128836similar to heterogeneous nuclear ribonucleoprotein A1 (217353_at), score: 0.78 LOC100129624hypothetical LOC100129624 (210748_at), score: 0.64 LOC100134401hypothetical protein LOC100134401 (213605_s_at), score: 0.6 LOC152719hypothetical protein LOC152719 (215978_x_at), score: 0.62 LOC171220destrin-2 pseudogene (211325_x_at), score: 0.65 LOC23117PI-3-kinase-related kinase SMG-1 isoform 1 homolog (211996_s_at), score: 0.66 LOC552889hypothetical protein LOC552889 (212114_at), score: 0.6 LOC643313similar to hypothetical protein LOC284701 (211050_x_at), score: 0.79 LOC644617hypothetical LOC644617 (221235_s_at), score: 0.77 LOC647070hypothetical LOC647070 (215467_x_at), score: 0.76 LOC728844hypothetical LOC728844 (222040_at), score: 0.65 MAGT1magnesium transporter 1 (210596_at), score: 0.64 MAP2K7mitogen-activated protein kinase kinase 7 (216206_x_at), score: 0.71 MAP3K9mitogen-activated protein kinase kinase kinase 9 (213927_at), score: 0.63 MCL1myeloid cell leukemia sequence 1 (BCL2-related) (214057_at), score: 0.86 MDN1MDN1, midasin homolog (yeast) (212693_at), score: 0.64 MEFVMediterranean fever (208262_x_at), score: 0.65 METAP2methionyl aminopeptidase 2 (202015_x_at), score: 0.8 METTL7Amethyltransferase like 7A (211424_x_at), score: 0.81 MFSD11major facilitator superfamily domain containing 11 (221192_x_at), score: 0.69 MLLT10myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 (216506_x_at), score: 0.7 MRPL44mitochondrial ribosomal protein L44 (218202_x_at), score: 0.69 MSL2male-specific lethal 2 homolog (Drosophila) (218733_at), score: 0.65 MXI1MAX interactor 1 (202364_at), score: 0.61 NACAnascent polypeptide-associated complex alpha subunit (222018_at), score: 0.8 NUP214nucleoporin 214kDa (202155_s_at), score: 0.65 OPHN1oligophrenin 1 (206323_x_at), score: 0.66 PBX2pre-B-cell leukemia homeobox 2 (202876_s_at), score: 0.8 PBXIP1pre-B-cell leukemia homeobox interacting protein 1 (214176_s_at), score: 0.69 PCGF2polycomb group ring finger 2 (203793_x_at), score: 0.59 PDE4Cphosphodiesterase 4C, cAMP-specific (phosphodiesterase E1 dunce homolog, Drosophila) (206792_x_at), score: 0.59 PELI2pellino homolog 2 (Drosophila) (219132_at), score: 0.87 PGFplacental growth factor (215179_x_at), score: 0.63 PHF21APHD finger protein 21A (203278_s_at), score: 0.62 PHF8PHD finger protein 8 (212916_at), score: 0.75 PLCL2phospholipase C-like 2 (213309_at), score: 0.69 POGZpogo transposable element with ZNF domain (215281_x_at), score: 0.78 POLR1Bpolymerase (RNA) I polypeptide B, 128kDa (220113_x_at), score: 0.73 POM121L9PPOM121 membrane glycoprotein-like 9 (rat) pseudogene (222253_s_at), score: 0.68 PRINSpsoriasis associated RNA induced by stress (non-protein coding) (216051_x_at), score: 0.78 PTGESprostaglandin E synthase (210367_s_at), score: 0.6 RAB11FIP1RAB11 family interacting protein 1 (class I) (219681_s_at), score: 0.76 RGL1ral guanine nucleotide dissociation stimulator-like 1 (209568_s_at), score: 0.66 RGS10regulator of G-protein signaling 10 (214000_s_at), score: 0.66 RGS5regulator of G-protein signaling 5 (218353_at), score: 0.9 RHEBRas homolog enriched in brain (213409_s_at), score: 0.6 RIPK2receptor-interacting serine-threonine kinase 2 (209545_s_at), score: 0.65 RNMTRNA (guanine-7-) methyltransferase (202683_s_at), score: 0.69 RPL14ribosomal protein L14 (219138_at), score: 0.6 RPL21P37ribosomal protein L21 pseudogene 37 (216479_at), score: 0.75 RPL23AP32ribosomal protein L23a pseudogene 32 (207283_at), score: 0.78 RPL35Aribosomal protein L35a (215208_x_at), score: 0.71 RPLP2ribosomal protein, large, P2 (200908_s_at), score: 0.68 RPS10P3ribosomal protein S10 pseudogene 3 (217336_at), score: 0.69 RPS19ribosomal protein S19 (202648_at), score: 0.71 RPS3AP47ribosomal protein S3a pseudogene 47 (216823_at), score: 0.77 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (214764_at), score: 0.67 SCARF1scavenger receptor class F, member 1 (206995_x_at), score: 0.63 SCD5stearoyl-CoA desaturase 5 (220232_at), score: 0.68 SCML2sex comb on midleg-like 2 (Drosophila) (206147_x_at), score: 0.64 SFRS11splicing factor, arginine/serine-rich 11 (213742_at), score: 0.72 SFRS18splicing factor, arginine/serine-rich 18 (212176_at), score: 0.65 SH3BP2SH3-domain binding protein 2 (217257_at), score: 0.85 SH3GL3SH3-domain GRB2-like 3 (211565_at), score: 0.87 SHOX2short stature homeobox 2 (210135_s_at), score: 0.6 SIAH1seven in absentia homolog 1 (Drosophila) (202981_x_at), score: 0.6 SKP1S-phase kinase-associated protein 1 (200719_at), score: 0.76 SLC11A1solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 (217473_x_at), score: 0.61 SLC17A6solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 (220551_at), score: 0.64 SLC30A5solute carrier family 30 (zinc transporter), member 5 (220181_x_at), score: 0.89 SLC6A6solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (205921_s_at), score: 0.65 SMEK2SMEK homolog 2, suppressor of mek1 (Dictyostelium) (222270_at), score: 0.71 SNX15sorting nexin 15 (205482_x_at), score: 0.61 SPATA2spermatogenesis associated 2 (204433_s_at), score: 0.62 SPG21spastic paraplegia 21 (autosomal recessive, Mast syndrome) (215383_x_at), score: 0.64 SPINLW1serine peptidase inhibitor-like, with Kunitz and WAP domains 1 (eppin) (206318_at), score: 0.86 SPNsialophorin (206057_x_at), score: 0.92 SPTLC3serine palmitoyltransferase, long chain base subunit 3 (220456_at), score: 0.87 STC1stanniocalcin 1 (204597_x_at), score: 0.61 SYCP1synaptonemal complex protein 1 (206740_x_at), score: 0.78 TCTN2tectonic family member 2 (206438_x_at), score: 0.73 TFAP2Ctranscription factor AP-2 gamma (activating enhancer binding protein 2 gamma) (205286_at), score: 0.64 TIMM8Atranslocase of inner mitochondrial membrane 8 homolog A (yeast) (210800_at), score: 0.9 TMEM38Btransmembrane protein 38B (218772_x_at), score: 0.67 TP53TG3TP53 target 3 (220167_s_at), score: 0.73 TRIM36tripartite motif-containing 36 (219736_at), score: 0.96 TRIT1tRNA isopentenyltransferase 1 (218617_at), score: 0.61 TRPV1transient receptor potential cation channel, subfamily V, member 1 (219632_s_at), score: 0.76 TULP4tubby like protein 4 (218184_at), score: 0.7 UBQLN4ubiquilin 4 (222252_x_at), score: 0.77 UGT2B28UDP glucuronosyltransferase 2 family, polypeptide B28 (211682_x_at), score: 0.75 UPP1uridine phosphorylase 1 (203234_at), score: 0.61 USP6ubiquitin specific peptidase 6 (Tre-2 oncogene) (206405_x_at), score: 0.65 VAMP2vesicle-associated membrane protein 2 (synaptobrevin 2) (201557_at), score: 0.77 VPS33Avacuolar protein sorting 33 homolog A (S. cerevisiae) (204590_x_at), score: 0.62 WDR43WD repeat domain 43 (214662_at), score: 0.59 XRCC2X-ray repair complementing defective repair in Chinese hamster cells 2 (207598_x_at), score: 0.72 ZBTB39zinc finger and BTB domain containing 39 (205256_at), score: 0.88 ZC3H7Bzinc finger CCCH-type containing 7B (206169_x_at), score: 0.8 ZEB2zinc finger E-box binding homeobox 2 (203603_s_at), score: 0.61 ZFC3H1zinc finger, C3H1-type containing (213065_at), score: 0.6 ZNF160zinc finger protein 160 (214715_x_at), score: 0.64 ZNF492zinc finger protein 492 (215532_x_at), score: 0.76 ZNF506zinc finger protein 506 (221626_at), score: 0.66 ZNF528zinc finger protein 528 (215019_x_at), score: 0.76 ZNF552zinc finger protein 552 (219741_x_at), score: 0.71 ZNF611zinc finger protein 611 (208137_x_at), score: 0.7 ZNF639zinc finger protein 639 (218413_s_at), score: 0.66 ZNF747zinc finger protein 747 (206180_x_at), score: 0.7 ZNF770zinc finger protein 770 (220608_s_at), score: 0.81 ZNF816Azinc finger protein 816A (217541_x_at), score: 0.84 ZSCAN12zinc finger and SCAN domain containing 12 (206507_at), score: 0.62
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |