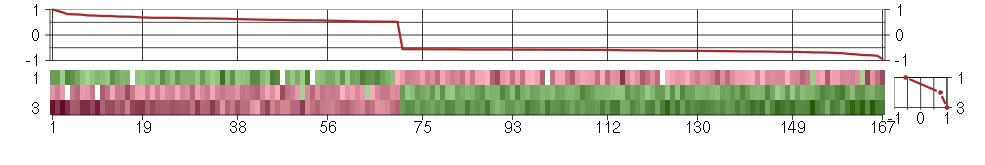

Under-expression is coded with green,
over-expression with red color.



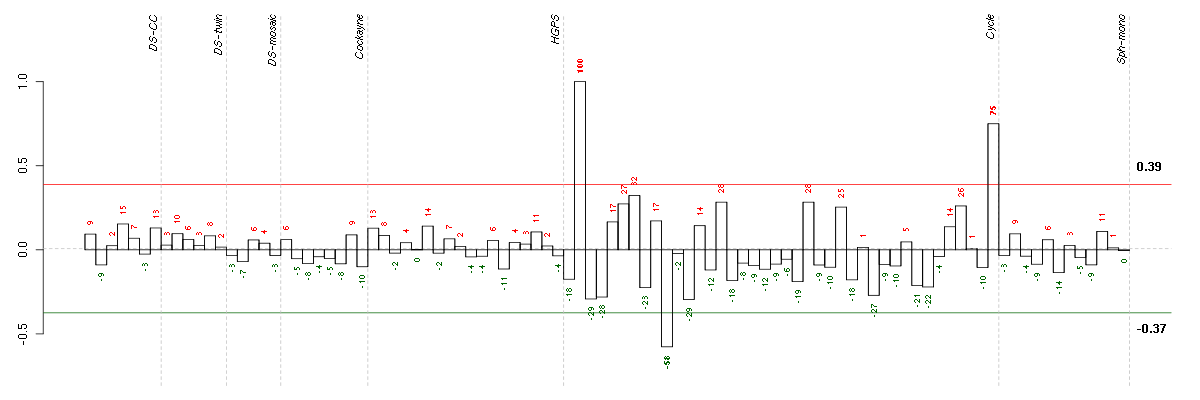
ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: 0.89 ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.6 ABOABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase) (216716_at), score: 0.53 ACSS3acyl-CoA synthetase short-chain family member 3 (219616_at), score: -0.56 ADAMTS3ADAM metallopeptidase with thrombospondin type 1 motif, 3 (214913_at), score: -0.57 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: 0.65 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: -0.56 ARHGAP24Rho GTPase activating protein 24 (221030_s_at), score: -0.56 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: 0.76 ATMataxia telangiectasia mutated (210858_x_at), score: 0.62 ATP6V0A1ATPase, H+ transporting, lysosomal V0 subunit a1 (205095_s_at), score: -0.59 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (214149_s_at), score: -0.58 ATP8B2ATPase, class I, type 8B, member 2 (216873_s_at), score: 0.54 B9D1B9 protein domain 1 (210534_s_at), score: -0.57 BANK1B-cell scaffold protein with ankyrin repeats 1 (219667_s_at), score: -0.61 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: 0.63 C11orf71chromosome 11 open reading frame 71 (218789_s_at), score: -0.59 C13orf23chromosome 13 open reading frame 23 (218420_s_at), score: -0.58 C20orf117chromosome 20 open reading frame 117 (207711_at), score: -0.66 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: 0.77 C2orf37chromosome 2 open reading frame 37 (220172_at), score: -0.62 CCDC40coiled-coil domain containing 40 (220592_at), score: 0.75 CCDC49coiled-coil domain containing 49 (218655_s_at), score: -0.58 CCDC9coiled-coil domain containing 9 (206257_at), score: 0.53 CCT6Bchaperonin containing TCP1, subunit 6B (zeta 2) (206587_at), score: -0.58 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.71 CEP164centrosomal protein 164kDa (204251_s_at), score: -0.8 CHRNA10cholinergic receptor, nicotinic, alpha 10 (220210_at), score: 0.59 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.57 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: -0.7 DDX28DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 (203785_s_at), score: -0.57 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: 0.67 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: 0.67 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: -0.82 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: -0.62 DUSP9dual specificity phosphatase 9 (205777_at), score: 0.61 DZIP3DAZ interacting protein 3, zinc finger (213186_at), score: -0.57 E2F5E2F transcription factor 5, p130-binding (221586_s_at), score: -0.56 ELL3elongation factor RNA polymerase II-like 3 (219517_at), score: -0.64 EPHA4EPH receptor A4 (206114_at), score: 0.56 EPHB4EPH receptor B4 (216680_s_at), score: 0.58 EPS8L1EPS8-like 1 (218778_x_at), score: 0.75 EXOC2exocyst complex component 2 (219349_s_at), score: -0.65 FAM105Afamily with sequence similarity 105, member A (219694_at), score: -0.64 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: -0.57 FGF13fibroblast growth factor 13 (205110_s_at), score: -0.58 FGF9fibroblast growth factor 9 (glia-activating factor) (206404_at), score: -0.6 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.94 FKRPfukutin related protein (219853_at), score: -0.64 FOXK2forkhead box K2 (203064_s_at), score: -0.62 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: -0.58 GZMMgranzyme M (lymphocyte met-ase 1) (207460_at), score: 0.59 HABP4hyaluronan binding protein 4 (209818_s_at), score: -0.56 HAMPhepcidin antimicrobial peptide (220491_at), score: 0.56 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: 0.53 HIST1H2AGhistone cluster 1, H2ag (207156_at), score: -0.64 HIST1H4Ghistone cluster 1, H4g (208551_at), score: 0.82 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: -0.68 HSPA6heat shock 70kDa protein 6 (HSP70B') (117_at), score: 0.65 HUNKhormonally up-regulated Neu-associated kinase (219535_at), score: 0.68 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: 0.79 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: 0.69 IRF2interferon regulatory factor 2 (203275_at), score: -0.74 JARID1Ajumonji, AT rich interactive domain 1A (215698_at), score: -0.62 KATNB1katanin p80 (WD repeat containing) subunit B 1 (203162_s_at), score: 0.67 KCNJ15potassium inwardly-rectifying channel, subfamily J, member 15 (210119_at), score: -0.58 KCTD17potassium channel tetramerisation domain containing 17 (205561_at), score: 0.58 KIAA0317KIAA0317 (202128_at), score: -0.56 KIAA1305KIAA1305 (220911_s_at), score: -0.79 KIF16Bkinesin family member 16B (219570_at), score: -0.62 KIR3DX1killer cell immunoglobulin-like receptor, three domains, X1 (216428_x_at), score: -0.56 KRT8P12keratin 8 pseudogene 12 (222060_at), score: 1 LILRA5leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 (215838_at), score: 0.52 LIMD1LIM domains containing 1 (218850_s_at), score: -0.62 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: 0.59 LOC149501similar to keratin 8 (216821_at), score: 0.67 LOC222070hypothetical protein LOC222070 (221949_at), score: -0.59 LOC644096hypothetical protein LOC644096 (221855_at), score: 0.52 LOC80054hypothetical LOC80054 (220465_at), score: 0.67 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: 0.63 LY6G5Clymphocyte antigen 6 complex, locus G5C (219860_at), score: -0.57 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: 0.74 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: 0.53 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: 0.57 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: -0.57 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.66 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: 0.6 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: -0.63 NEFMneurofilament, medium polypeptide (205113_at), score: 0.81 NEO1neogenin homolog 1 (chicken) (204321_at), score: -0.67 NF1neurofibromin 1 (211094_s_at), score: -0.61 NFYCnuclear transcription factor Y, gamma (202215_s_at), score: -0.63 NME5non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase) (206197_at), score: -0.56 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: 0.66 NPEPL1aminopeptidase-like 1 (89476_r_at), score: 0.64 NUPL2nucleoporin like 2 (204003_s_at), score: 0.55 NXPH3neurexophilin 3 (221991_at), score: 0.65 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: 0.58 P2RX5purinergic receptor P2X, ligand-gated ion channel, 5 (210448_s_at), score: 0.57 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: -0.66 PAIP1poly(A) binding protein interacting protein 1 (208051_s_at), score: -0.56 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: -0.55 PDPRpyruvate dehydrogenase phosphatase regulatory subunit (220236_at), score: -0.63 PECRperoxisomal trans-2-enoyl-CoA reductase (221142_s_at), score: -0.67 PGK2phosphoglycerate kinase 2 (217009_at), score: 0.6 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: -0.76 PHF2PHD finger protein 2 (212726_at), score: -0.79 PIAS3protein inhibitor of activated STAT, 3 (203035_s_at), score: -0.66 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: -0.65 PLA2G4Cphospholipase A2, group IVC (cytosolic, calcium-independent) (209785_s_at), score: -0.58 PLAC8placenta-specific 8 (219014_at), score: -0.58 POLR3Fpolymerase (RNA) III (DNA directed) polypeptide F, 39 kDa (205218_at), score: -0.58 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: 0.72 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: 0.52 PTPN20Bprotein tyrosine phosphatase, non-receptor type 20B (215172_at), score: -0.56 RAB11BRAB11B, member RAS oncogene family (34478_at), score: -0.58 RBM12BRNA binding motif protein 12B (51228_at), score: -0.65 RBM3RNA binding motif (RNP1, RRM) protein 3 (222026_at), score: 0.54 RBM38RNA binding motif protein 38 (212430_at), score: 0.72 RBM4BRNA binding motif protein 4B (209497_s_at), score: -0.58 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: -0.58 ROS1c-ros oncogene 1 , receptor tyrosine kinase (207569_at), score: 0.64 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.59 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (203379_at), score: 0.77 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.62 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: -0.65 SETD1ASET domain containing 1A (213202_at), score: 0.72 SF3A2splicing factor 3a, subunit 2, 66kDa (37462_i_at), score: -0.57 SH2D3ASH2 domain containing 3A (222169_x_at), score: 0.65 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: -0.69 SIX6SIX homeobox 6 (207250_at), score: 0.67 SMA4glucuronidase, beta pseudogene (215599_at), score: -0.62 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: -0.71 SMYD5SMYD family member 5 (209516_at), score: -0.57 SNRKSNF related kinase (209481_at), score: -0.58 SNX27sorting nexin family member 27 (221006_s_at), score: -0.61 STIM1stromal interaction molecule 1 (202764_at), score: -0.6 STX17syntaxin 17 (218666_s_at), score: -0.59 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: 0.57 TCP10t-complex 10 homolog (mouse) (207503_at), score: 0.81 TMPRSS6transmembrane protease, serine 6 (214955_at), score: 0.7 TNFSF10tumor necrosis factor (ligand) superfamily, member 10 (214329_x_at), score: 0.64 TP53tumor protein p53 (201746_at), score: -0.62 TRA@T cell receptor alpha locus (216540_at), score: 0.58 TRIM21tripartite motif-containing 21 (204804_at), score: -0.64 TRIM45tripartite motif-containing 45 (219923_at), score: 0.55 TRY6trypsinogen C (215395_x_at), score: 0.52 TSKStestis-specific serine kinase substrate (220545_s_at), score: 0.6 TSKUtsukushin (218245_at), score: -0.6 TTC30Atetratricopeptide repeat domain 30A (213679_at), score: -0.94 TWISTNBTWIST neighbor (214729_at), score: -0.58 UGT2A1UDP glucuronosyltransferase 2 family, polypeptide A1 (207958_at), score: -0.69 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: -0.68 WFDC8WAP four-disulfide core domain 8 (215276_at), score: 0.67 WHSC2Wolf-Hirschhorn syndrome candidate 2 (34225_at), score: -0.62 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: 0.52 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: 0.61 ZBTB6zinc finger and BTB domain containing 6 (206098_at), score: -0.66 ZNF132zinc finger protein 132 (207402_at), score: -0.59 ZNF141zinc finger protein 141 (206931_at), score: -0.57 ZNF192zinc finger protein 192 (206579_at), score: -0.64 ZNF3zinc finger protein 3 (212684_at), score: -0.75 ZNF304zinc finger protein 304 (207753_at), score: -0.56 ZNF365zinc finger protein 365 (206448_at), score: -0.58 ZNF434zinc finger protein 434 (218937_at), score: -0.66 ZNF518Azinc finger protein 518A (204291_at), score: -0.71 ZNF814zinc finger protein 814 (60794_f_at), score: -0.6
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |