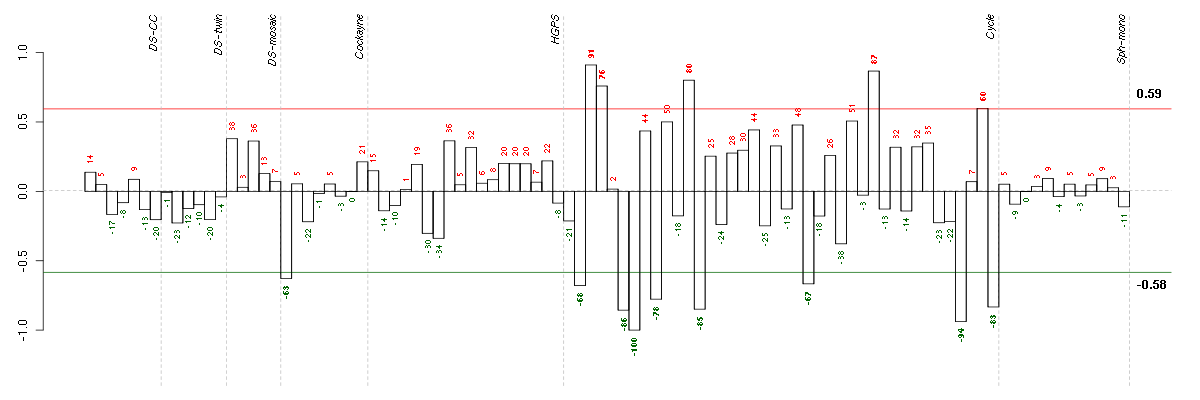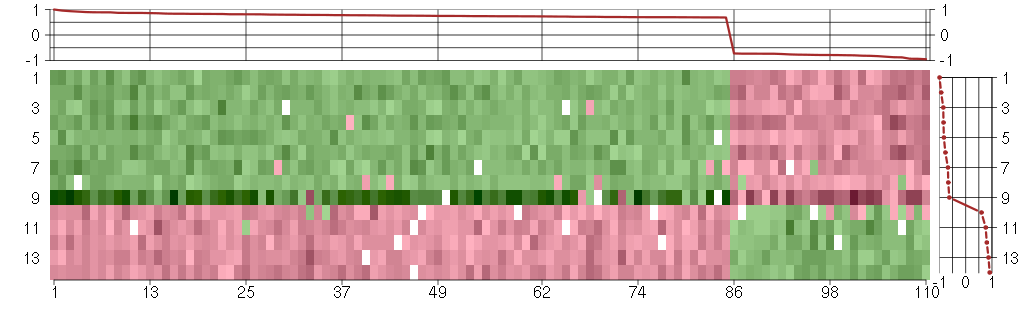

Under-expression is coded with green,
over-expression with red color.

biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
positive regulation of catalytic activity
Any process that activates or increases the activity of an enzyme.
regulation of MAP kinase activity
Any process that modulates the frequency, rate or extent of MAP kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
regulation of JUN kinase activity
Any process that modulates the frequency, rate or extent of JUN kinase activity.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.
regulation of kinase activity
Any process that modulates the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
regulation of protein kinase activity
Any process that modulates the frequency, rate or extent of protein kinase activity.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of transferase activity
Any process that modulates the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of molecular functions. Molecular functions are elemental biological activities occurring at the molecular level, such as catalysis or binding.
all
This term is the most general term possible
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.


ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.74 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.73 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.74 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.8 ARSAarylsulfatase A (204443_at), score: 0.69 ATF5activating transcription factor 5 (204999_s_at), score: 0.69 ATN1atrophin 1 (40489_at), score: 0.74 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.73 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.8 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.87 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.74 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.79 CALB2calbindin 2 (205428_s_at), score: -0.93 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.72 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.89 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: 0.73 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: 0.83 CICcapicua homolog (Drosophila) (212784_at), score: 0.87 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.8 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.81 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.87 CScitrate synthase (208660_at), score: 0.75 DENND3DENN/MADD domain containing 3 (212974_at), score: 0.69 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.81 EPHA4EPH receptor A4 (206114_at), score: -0.81 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.92 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.69 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.75 FOXK2forkhead box K2 (203064_s_at), score: 0.96 GMPRguanosine monophosphate reductase (204187_at), score: 0.77 H1FXH1 histone family, member X (204805_s_at), score: 0.8 HAB1B1 for mucin (215778_x_at), score: -0.82 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.84 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: 0.71 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.85 HSF1heat shock transcription factor 1 (202344_at), score: 0.7 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.78 IL9Rinterleukin 9 receptor (217212_s_at), score: -0.79 INTS1integrator complex subunit 1 (212212_s_at), score: 0.7 JUNDjun D proto-oncogene (203751_x_at), score: 0.77 JUPjunction plakoglobin (201015_s_at), score: 0.93 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.88 KIAA1305KIAA1305 (220911_s_at), score: 0.76 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.79 LEPREL2leprecan-like 2 (204854_at), score: 0.73 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.8 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.87 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.83 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.73 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.85 NEFMneurofilament, medium polypeptide (205113_at), score: -0.95 NF1neurofibromin 1 (211094_s_at), score: 0.7 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.7 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.79 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.72 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.71 PARVBparvin, beta (37965_at), score: 0.82 PCDH9protocadherin 9 (219737_s_at), score: -0.87 PCNXpecanex homolog (Drosophila) (213159_at), score: 0.73 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.71 PHF7PHD finger protein 7 (215622_x_at), score: -0.76 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.83 PKN1protein kinase N1 (202161_at), score: 0.76 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.76 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.76 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.7 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.7 PRKCDprotein kinase C, delta (202545_at), score: 0.77 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.75 PXNpaxillin (211823_s_at), score: 0.79 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.75 RBM15BRNA binding motif protein 15B (202689_at), score: 0.7 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.89 RNF220ring finger protein 220 (219988_s_at), score: 0.71 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: -0.73 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.93 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.7 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.77 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.79 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.77 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 1 SF1splicing factor 1 (208313_s_at), score: 0.86 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.72 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.78 SMAD3SMAD family member 3 (218284_at), score: 0.75 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.78 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.71 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.83 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.78 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: -0.79 TAF13TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa (205966_at), score: -0.74 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.81 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.69 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.69 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.73 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.73 TLE3transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) (206472_s_at), score: 0.73 TLE4transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) (204872_at), score: -0.77 TP53tumor protein p53 (201746_at), score: 0.83 TRIM21tripartite motif-containing 21 (204804_at), score: 0.76 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.73 TSKUtsukushin (218245_at), score: 0.82 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.78 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.9 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.81 WDR42AWD repeat domain 42A (202249_s_at), score: 0.81 WDR6WD repeat domain 6 (217734_s_at), score: 0.89 XAB2XPA binding protein 2 (218110_at), score: 0.72 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.75
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |