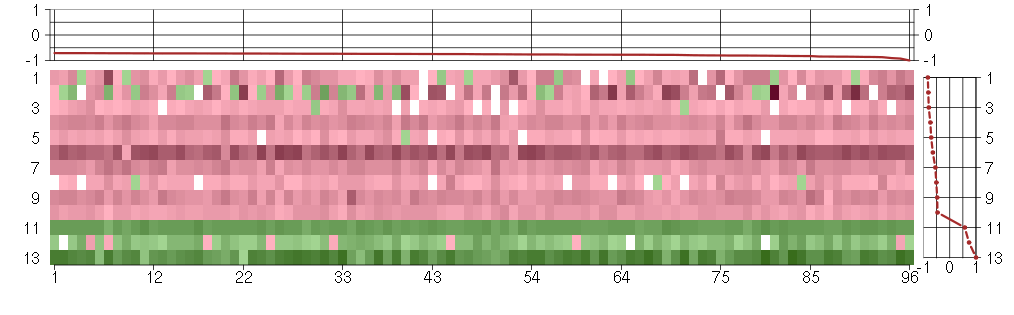

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism.
ion transport
The directed movement of charged atoms or small charged molecules into, out of, within or between cells.
anion transport
The directed movement of anions, atoms or small molecules with a net negative charge, into, out of, within or between cells.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
sensory perception
The series of events required for an organism to receive a sensory stimulus, convert it to a molecular signal, and recognize and characterize the signal.
cell recognition
The process by which a cell in a multicellular organism interprets its surroundings.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
organic anion transport
The directed movement of organic anions into, out of, within or between cells. Organic anions are atoms or small molecules with a negative charge which contain carbon in covalent linkage.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
cognition
The operation of the mind by which an organism becomes aware of objects of thought or perception; it includes the mental activities associated with thinking, learning, and memory.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
all
This term is the most general term possible
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.

membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
anchored to membrane
Tethered to a membrane by a covalently attached anchor, such as a lipid moiety, that is embedded in the membrane. When used to describe a protein, indicates that none of the peptide sequence is embedded in the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
transmembrane transporter activity
Catalysis of the transfer of a substance from one side of a membrane to the other.
organic acid transmembrane transporter activity
Catalysis of the transfer of organic acids, any acidic compound containing carbon in covalent linkage, from one side of the membrane to the other.
monocarboxylic acid transmembrane transporter activity
Catalysis of the transfer of monocarboxylic acids from one side of the membrane to the other. A monocarboxylic acid is an organic acid with one COOH group.
bile acid transmembrane transporter activity
Catalysis of the transfer of bile acid from one side of the membrane to the other. Bile acids are any of a group of steroid carboxylic acids occurring in bile, where they are present as the sodium salts of their amides with glycine or taurine.
substrate-specific transmembrane transporter activity
Catalysis of the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
carboxylic acid transmembrane transporter activity
Catalysis of the transfer of carboxylic acids from one side of the membrane to the other. Carboxylic acids are organic acids containing one or more carboxyl (COOH) groups or anions (COO-).
all
This term is the most general term possible
substrate-specific transmembrane transporter activity
Catalysis of the transfer of a specific substance or group of related substances from one side of a membrane to the other.
ADAMTS9ADAM metallopeptidase with thrombospondin type 1 motif, 9 (220287_at), score: -0.71 ADAMTSL4ADAMTS-like 4 (220578_at), score: -0.74 AFF2AF4/FMR2 family, member 2 (206105_at), score: -0.81 AGBL2ATP/GTP binding protein-like 2 (220390_at), score: -0.75 AGMATagmatine ureohydrolase (agmatinase) (219792_at), score: -0.74 APOMapolipoprotein M (205682_x_at), score: -0.77 ATRNL1attractin-like 1 (213744_at), score: -0.85 BEND5BEN domain containing 5 (219670_at), score: -0.74 BMP10bone morphogenetic protein 10 (208292_at), score: -0.73 BTNL3butyrophilin-like 3 (217207_s_at), score: -0.74 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: -0.79 C8orf17chromosome 8 open reading frame 17 (208266_at), score: -0.72 CA1carbonic anhydrase I (205949_at), score: -0.85 CA4carbonic anhydrase IV (206209_s_at), score: -0.73 CARTPTCART prepropeptide (206339_at), score: -0.72 CD53CD53 molecule (203416_at), score: -0.86 CEACAM5carcinoembryonic antigen-related cell adhesion molecule 5 (201884_at), score: -0.75 CEACAM6carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) (211657_at), score: -0.74 CHD2chromodomain helicase DNA binding protein 2 (203461_at), score: -0.72 CLDN10claudin 10 (205328_at), score: -0.87 CLEC4MC-type lectin domain family 4, member M (207995_s_at), score: -0.82 CLEC7AC-type lectin domain family 7, member A (221698_s_at), score: -0.72 CNKSR2connector enhancer of kinase suppressor of Ras 2 (206731_at), score: -0.85 CTRLchymotrypsin-like (214377_s_at), score: -0.77 CYP2C9cytochrome P450, family 2, subfamily C, polypeptide 9 (214421_x_at), score: -0.71 CYP2E1cytochrome P450, family 2, subfamily E, polypeptide 1 (1431_at), score: -0.76 DAOD-amino-acid oxidase (206878_at), score: -0.8 DNASE1L3deoxyribonuclease I-like 3 (205554_s_at), score: -0.73 DRD5dopamine receptor D5 (208486_at), score: -0.73 ELNelastin (212670_at), score: -0.75 FAM38Bfamily with sequence similarity 38, member B (219602_s_at), score: -0.8 GABRR2gamma-aminobutyric acid (GABA) receptor, rho 2 (208217_at), score: -0.71 GPATCH4G patch domain containing 4 (220596_at), score: -0.73 GRHL2grainyhead-like 2 (Drosophila) (219388_at), score: -0.8 HOXD4homeobox D4 (205522_at), score: -0.74 IFNA7interferon, alpha 7 (208259_x_at), score: -0.71 IL16interleukin 16 (lymphocyte chemoattractant factor) (209827_s_at), score: -0.77 IL1RAPL1interleukin 1 receptor accessory protein-like 1 (220663_at), score: -0.72 ITIH2inter-alpha (globulin) inhibitor H2 (204987_at), score: -0.76 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: -0.72 KIR3DX1killer cell immunoglobulin-like receptor, three domains, X1 (216428_x_at), score: -0.76 KLF1Kruppel-like factor 1 (erythroid) (210504_at), score: -0.73 KNG1kininogen 1 (217512_at), score: -0.76 KRT2keratin 2 (207908_at), score: -0.91 LHX3LIM homeobox 3 (221670_s_at), score: -0.83 LOC100188945cell division cycle associated 4 pseudogene (215109_at), score: -0.77 LOC389906similar to Serine/threonine-protein kinase PRKX (Protein kinase PKX1) (59433_at), score: -0.73 LOC730227hypothetical protein LOC730227 (215756_at), score: -0.73 LOC93432maltase-glucoamylase-like pseudogene (216666_at), score: -0.76 LONRF3LON peptidase N-terminal domain and ring finger 3 (220009_at), score: -0.81 LPAR4lysophosphatidic acid receptor 4 (206960_at), score: -0.76 MBmyoglobin (204179_at), score: -0.85 MCF2L2MCF.2 cell line derived transforming sequence-like 2 (215112_x_at), score: -0.72 MIA2melanoma inhibitory activity 2 (221177_at), score: -0.75 MMRN2multimerin 2 (219091_s_at), score: -0.73 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: -0.85 N4BP3Nedd4 binding protein 3 (214775_at), score: -0.74 NINninein (GSK3B interacting protein) (219285_s_at), score: -0.74 NR0B1nuclear receptor subfamily 0, group B, member 1 (206644_at), score: -0.73 NRN1neuritin 1 (218625_at), score: -0.81 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: -0.79 OR2C1olfactory receptor, family 2, subfamily C, member 1 (221460_at), score: -0.72 OR7E24olfactory receptor, family 7, subfamily E, member 24 (215463_at), score: -0.81 PAX4paired box 4 (207867_at), score: -0.73 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: -0.86 PEG3paternally expressed 3 (209242_at), score: -0.83 PGCprogastricsin (pepsinogen C) (205261_at), score: -0.81 PMP2peripheral myelin protein 2 (206826_at), score: -0.77 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: -0.76 POU4F1POU class 4 homeobox 1 (206940_s_at), score: -0.72 PPP4R4protein phosphatase 4, regulatory subunit 4 (220672_at), score: -0.71 PROL1proline rich, lacrimal 1 (208004_at), score: -0.76 PROX1prospero homeobox 1 (207401_at), score: -0.73 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: -0.77 RAPGEF4Rap guanine nucleotide exchange factor (GEF) 4 (205651_x_at), score: -0.78 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: -1 RRHretinal pigment epithelium-derived rhodopsin homolog (208314_at), score: -0.77 S100A14S100 calcium binding protein A14 (218677_at), score: -0.72 SIRPB1signal-regulatory protein beta 1 (206934_at), score: -0.78 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: -0.71 SLC10A2solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (207095_at), score: -0.78 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: -0.75 SLC26A4solute carrier family 26, member 4 (206529_x_at), score: -0.77 SLC4A5solute carrier family 4, sodium bicarbonate cotransporter, member 5 (204296_at), score: -0.73 SLCO1A2solute carrier organic anion transporter family, member 1A2 (207308_at), score: -0.74 SPAG6sperm associated antigen 6 (210033_s_at), score: -0.72 SPAM1sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding) (216989_at), score: -0.74 TACR1tachykinin receptor 1 (208048_at), score: -0.92 TAS2R16taste receptor, type 2, member 16 (221444_at), score: -0.72 TBX21T-box 21 (220684_at), score: -0.78 TMEM19transmembrane protein 19 (219941_at), score: -0.77 TP63tumor protein p63 (209863_s_at), score: -0.82 TRA@T cell receptor alpha locus (216540_at), score: -0.76 VGFVGF nerve growth factor inducible (205586_x_at), score: -0.72 ZNF335zinc finger protein 335 (78330_at), score: -0.75
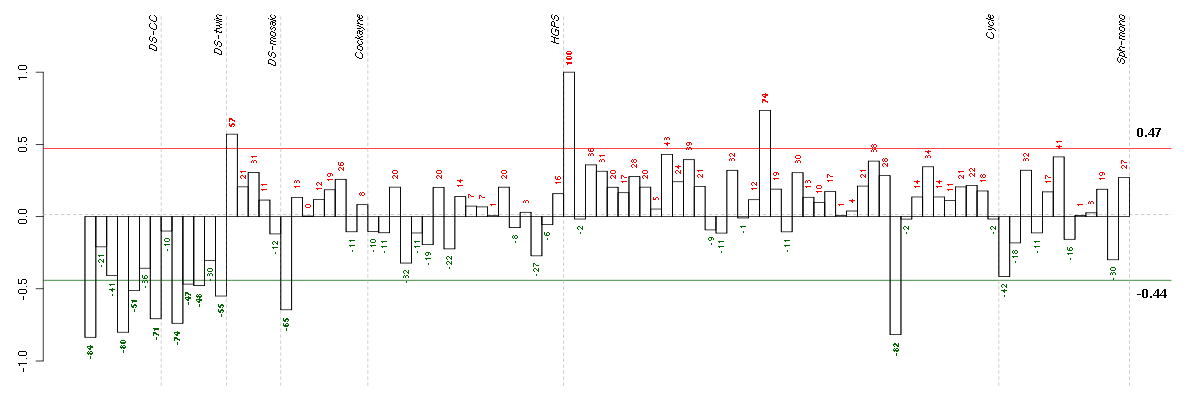
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |