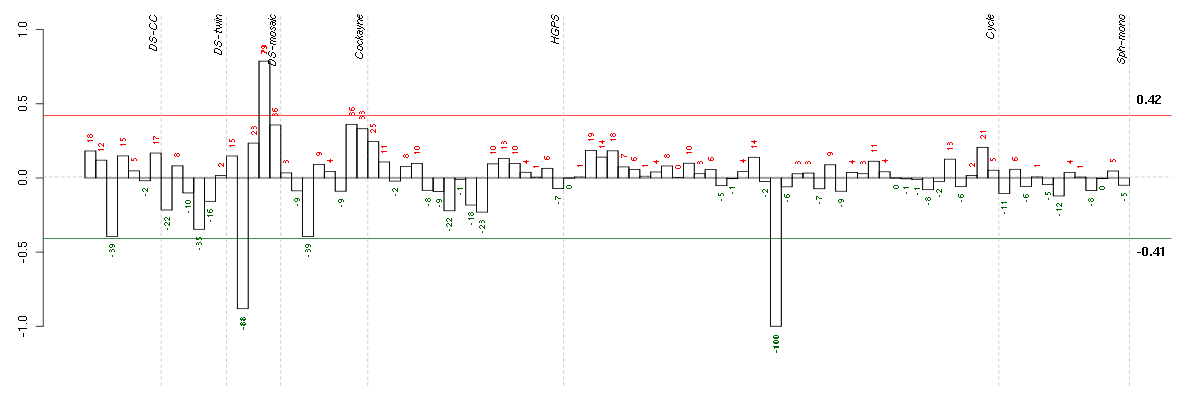

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04060 | 1.612e-02 | 4.193 | 13 | 115 | Cytokine-cytokine receptor interaction |
ABCC5ATP-binding cassette, sub-family C (CFTR/MRP), member 5 (209380_s_at), score: -0.53 ADAM15ADAM metallopeptidase domain 15 (217007_s_at), score: 0.81 ADAMTS12ADAM metallopeptidase with thrombospondin type 1 motif, 12 (221421_s_at), score: 0.51 ADAMTS2ADAM metallopeptidase with thrombospondin type 1 motif, 2 (214454_at), score: 0.42 ADRBK1adrenergic, beta, receptor kinase 1 (201401_s_at), score: 0.7 AFTPHaftiphilin (217939_s_at), score: -0.53 AGPAT31-acylglycerol-3-phosphate O-acyltransferase 3 (219723_x_at), score: 0.39 ALDH3B1aldehyde dehydrogenase 3 family, member B1 (211004_s_at), score: 0.42 ALDH4A1aldehyde dehydrogenase 4 family, member A1 (211552_s_at), score: 0.44 AP1S1adaptor-related protein complex 1, sigma 1 subunit (205195_at), score: 0.52 APBA1amyloid beta (A4) precursor protein-binding, family A, member 1 (206679_at), score: 0.59 APPL1adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 (218158_s_at), score: -0.56 ARFGAP2ADP-ribosylation factor GTPase activating protein 2 (211975_at), score: -0.43 ARFRP1ADP-ribosylation factor related protein 1 (215984_s_at), score: 0.4 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 0.59 ARL17P1ADP-ribosylation factor-like 17 pseudogene 1 (210718_s_at), score: -0.46 ARPC4actin related protein 2/3 complex, subunit 4, 20kDa (211672_s_at), score: 0.46 ASH1Lash1 (absent, small, or homeotic)-like (Drosophila) (218554_s_at), score: 0.39 ASPHaspartate beta-hydroxylase (205808_at), score: 0.41 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: 0.41 ATP13A3ATPase type 13A3 (219558_at), score: 0.62 ATP6V0A1ATPase, H+ transporting, lysosomal V0 subunit a1 (205095_s_at), score: 0.39 ATP6V1AATPase, H+ transporting, lysosomal 70kDa, V1 subunit A (201971_s_at), score: 0.45 ATP7AATPase, Cu++ transporting, alpha polypeptide (205197_s_at), score: 0.43 BAXBCL2-associated X protein (208478_s_at), score: 0.43 BCAT1branched chain aminotransferase 1, cytosolic (214452_at), score: 0.41 BCHEbutyrylcholinesterase (205433_at), score: 0.54 BCL2L1BCL2-like 1 (215037_s_at), score: 0.69 BET1Lblocked early in transport 1 homolog (S. cerevisiae)-like (220470_at), score: 0.62 BNC2basonuclin 2 (220272_at), score: -0.43 BSGbasigin (Ok blood group) (208677_s_at), score: 0.44 C14orf118chromosome 14 open reading frame 118 (219720_s_at), score: 0.43 C14orf133chromosome 14 open reading frame 133 (218431_at), score: -0.43 C19orf22chromosome 19 open reading frame 22 (221764_at), score: 0.52 C19orf6chromosome 19 open reading frame 6 (213986_s_at), score: 1 CACNA2D1calcium channel, voltage-dependent, alpha 2/delta subunit 1 (207050_at), score: 0.53 CAMK2Bcalcium/calmodulin-dependent protein kinase II beta (34846_at), score: 0.47 CCL7chemokine (C-C motif) ligand 7 (208075_s_at), score: 0.46 CD44CD44 molecule (Indian blood group) (210916_s_at), score: 0.52 CD82CD82 molecule (203904_x_at), score: 0.45 CDK6cyclin-dependent kinase 6 (207143_at), score: 0.59 CDV3CDV3 homolog (mouse) (213548_s_at), score: 0.52 CFIcomplement factor I (203854_at), score: -0.48 CHKAcholine kinase alpha (204233_s_at), score: 0.54 CHMchoroideremia (Rab escort protein 1) (207099_s_at), score: 0.4 CHRM2cholinergic receptor, muscarinic 2 (221330_at), score: 0.44 CHST5carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 (219182_at), score: -0.51 CHST7carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 (206756_at), score: -0.45 CLPTM1cleft lip and palate associated transmembrane protein 1 (211136_s_at), score: 0.53 CLTCL1clathrin, heavy chain-like 1 (205944_s_at), score: -0.6 CNOT4CCR4-NOT transcription complex, subunit 4 (203291_at), score: -0.47 COL1A1collagen, type I, alpha 1 (217430_x_at), score: 0.45 COL6A1collagen, type VI, alpha 1 (212940_at), score: 0.77 COPAcoatomer protein complex, subunit alpha (214336_s_at), score: 0.88 CPDcarboxypeptidase D (201942_s_at), score: 0.67 CTAGE5CTAGE family, member 5 (215930_s_at), score: 0.49 CYTH3cytohesin 3 (206523_at), score: 0.51 CYTIPcytohesin 1 interacting protein (209606_at), score: 0.44 DAPK3death-associated protein kinase 3 (203890_s_at), score: 0.59 DCTN5dynactin 5 (p25) (209231_s_at), score: 0.4 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: 0.64 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: 0.66 DISC1disrupted in schizophrenia 1 (206090_s_at), score: -0.62 DLG4discs, large homolog 4 (Drosophila) (204592_at), score: 0.43 DSC3desmocollin 3 (206032_at), score: 0.44 E2F5E2F transcription factor 5, p130-binding (221586_s_at), score: -0.5 EDC3enhancer of mRNA decapping 3 homolog (S. cerevisiae) (219207_at), score: 0.43 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: -0.53 EHMT1euchromatic histone-lysine N-methyltransferase 1 (219339_s_at), score: 0.73 EIF5Aeukaryotic translation initiation factor 5A (201123_s_at), score: 0.49 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: 0.91 ENSAendosulfine alpha (221487_s_at), score: 0.41 ENTPD6ectonucleoside triphosphate diphosphohydrolase 6 (putative function) (201704_at), score: -0.44 EPRSglutamyl-prolyl-tRNA synthetase (200841_s_at), score: 0.39 ETS1v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) (214447_at), score: 0.42 ETV7ets variant 7 (221680_s_at), score: 0.39 EXTL3exostoses (multiple)-like 3 (211051_s_at), score: 0.46 EZRezrin (208621_s_at), score: 0.44 FAM118Afamily with sequence similarity 118, member A (219629_at), score: -0.48 FGD1FYVE, RhoGEF and PH domain containing 1 (204819_at), score: 0.39 FGFR1fibroblast growth factor receptor 1 (210973_s_at), score: 0.52 FKBP15FK506 binding protein 15, 133kDa (76897_s_at), score: 0.45 FN1fibronectin 1 (214701_s_at), score: 0.86 FNTBfarnesyltransferase, CAAX box, beta (204764_at), score: 0.52 FRMD4AFERM domain containing 4A (208476_s_at), score: 0.54 FSCN1fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) (210933_s_at), score: 0.48 GATCglutamyl-tRNA(Gln) amidotransferase, subunit C homolog (bacterial) (214711_at), score: 0.67 GCAgrancalcin, EF-hand calcium binding protein (203765_at), score: -0.5 GGA2golgi associated, gamma adaptin ear containing, ARF binding protein 2 (210658_s_at), score: 0.4 GLULglutamate-ammonia ligase (glutamine synthetase) (200648_s_at), score: 0.5 GNRHRgonadotropin-releasing hormone receptor (216341_s_at), score: 0.41 GNSglucosamine (N-acetyl)-6-sulfatase (203676_at), score: 0.46 GP1BAglycoprotein Ib (platelet), alpha polypeptide (207389_at), score: 0.39 GPR137G protein-coupled receptor 137 (43934_at), score: 0.55 GPR176G protein-coupled receptor 176 (206673_at), score: 0.4 GPR56G protein-coupled receptor 56 (212070_at), score: -0.48 GTF2Igeneral transcription factor II, i (210892_s_at), score: 0.55 HECW1HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 (215584_at), score: 0.54 HIST1H2BGhistone cluster 1, H2bg (210387_at), score: 0.4 HIST1H2BIhistone cluster 1, H2bi (208523_x_at), score: 0.47 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.51 HP1BP3heterochromatin protein 1, binding protein 3 (220633_s_at), score: 0.58 IFNA16interferon, alpha 16 (208448_x_at), score: 0.43 IFNAR1interferon (alpha, beta and omega) receptor 1 (204191_at), score: 0.51 IGFBP5insulin-like growth factor binding protein 5 (203424_s_at), score: 0.41 IL17RCinterleukin 17 receptor C (64440_at), score: 0.66 KBTBD2kelch repeat and BTB (POZ) domain containing 2 (212447_at), score: -0.44 KCNAB1potassium voltage-gated channel, shaker-related subfamily, beta member 1 (210078_s_at), score: 0.45 KCTD20potassium channel tetramerisation domain containing 20 (214849_at), score: 0.81 KIAA0556KIAA0556 (213185_at), score: -0.43 KITLGKIT ligand (211124_s_at), score: 0.53 KLF7Kruppel-like factor 7 (ubiquitous) (204334_at), score: 0.42 LAMP1lysosomal-associated membrane protein 1 (201551_s_at), score: 0.63 LIMD1LIM domains containing 1 (218850_s_at), score: 0.49 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: 0.59 LOC222070hypothetical protein LOC222070 (221949_at), score: -0.43 LOC285412similar to KIAA0737 protein (217448_s_at), score: 0.59 LOC730092RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene (214712_at), score: -0.52 LOC730092RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene (216908_x_at), score: 0.57 LOXlysyl oxidase (213640_s_at), score: 0.53 LOXL2lysyl oxidase-like 2 (202997_s_at), score: 0.61 LRP12low density lipoprotein-related protein 12 (220253_s_at), score: 0.44 LRRN2leucine rich repeat neuronal 2 (216164_at), score: 0.55 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (211019_s_at), score: 0.49 MADCAM1mucosal vascular addressin cell adhesion molecule 1 (208037_s_at), score: 0.41 MADDMAP-kinase activating death domain (38398_at), score: -0.58 MAFKv-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian) (206750_at), score: 0.72 MAPKAPK2mitogen-activated protein kinase-activated protein kinase 2 (201461_s_at), score: 0.42 MAPRE3microtubule-associated protein, RP/EB family, member 3 (214270_s_at), score: 0.55 MARCH6membrane-associated ring finger (C3HC4) 6 (201737_s_at), score: 0.47 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: -0.43 MAXMYC associated factor X (210734_x_at), score: 0.66 MAZMYC-associated zinc finger protein (purine-binding transcription factor) (207824_s_at), score: 0.53 MBTPS2membrane-bound transcription factor peptidase, site 2 (206473_at), score: 0.39 MC1Rmelanocortin 1 receptor (alpha melanocyte stimulating hormone receptor) (205458_at), score: 0.39 MDM2Mdm2 p53 binding protein homolog (mouse) (217373_x_at), score: 0.47 METmet proto-oncogene (hepatocyte growth factor receptor) (211599_x_at), score: 0.4 METT11D1methyltransferase 11 domain containing 1 (218366_x_at), score: -0.46 MFN2mitofusin 2 (216205_s_at), score: 0.6 MGC31957hypothetical protein MGC31957 (210484_s_at), score: 0.45 MMP14matrix metallopeptidase 14 (membrane-inserted) (202827_s_at), score: 0.61 MPDU1mannose-P-dolichol utilization defect 1 (209208_at), score: 0.43 MRASmuscle RAS oncogene homolog (206538_at), score: 0.4 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: 0.43 MYST3MYST histone acetyltransferase (monocytic leukemia) 3 (202423_at), score: -0.48 NAALAD2N-acetylated alpha-linked acidic dipeptidase 2 (222161_at), score: -0.54 NAB2NGFI-A binding protein 2 (EGR1 binding protein 2) (216017_s_at), score: 0.56 NFE2L3nuclear factor (erythroid-derived 2)-like 3 (204702_s_at), score: 0.55 NPAL2NIPA-like domain containing 2 (220128_s_at), score: 0.46 NPTX1neuronal pentraxin I (204684_at), score: 0.47 NR1H3nuclear receptor subfamily 1, group H, member 3 (203920_at), score: -0.47 NUMA1nuclear mitotic apparatus protein 1 (214251_s_at), score: 0.63 OR1E1olfactory receptor, family 1, subfamily E, member 1 (214515_at), score: 0.4 OR2C1olfactory receptor, family 2, subfamily C, member 1 (221460_at), score: 0.39 PAFAH1B1platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa (211547_s_at), score: 0.56 PARP8poly (ADP-ribose) polymerase family, member 8 (219033_at), score: -0.51 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.8 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.85 PCDHGB5protocadherin gamma subfamily B, 5 (211807_x_at), score: 0.43 PCF11PCF11, cleavage and polyadenylation factor subunit, homolog (S. cerevisiae) (203378_at), score: -0.64 PCIF1PDX1 C-terminal inhibiting factor 1 (222045_s_at), score: 0.42 PDZRN4PDZ domain containing ring finger 4 (220595_at), score: -0.58 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: 0.4 PHF15PHD finger protein 15 (212660_at), score: -0.45 PHLPPLPH domain and leucine rich repeat protein phosphatase-like (213407_at), score: -0.51 PHTF2putative homeodomain transcription factor 2 (217097_s_at), score: 0.58 PIAS3protein inhibitor of activated STAT, 3 (203035_s_at), score: -0.48 PIONpigeon homolog (Drosophila) (222150_s_at), score: -0.7 PIP5K1Aphosphatidylinositol-4-phosphate 5-kinase, type I, alpha (211205_x_at), score: 0.46 PISDphosphatidylserine decarboxylase (202392_s_at), score: -0.44 PLAGL2pleiomorphic adenoma gene-like 2 (202925_s_at), score: -0.44 PLEC1plectin 1, intermediate filament binding protein 500kDa (216971_s_at), score: 0.71 PLXNB3plexin B3 (205957_at), score: 0.41 POLDIP3polymerase (DNA-directed), delta interacting protein 3 (215357_s_at), score: 0.71 POLSpolymerase (DNA directed) sigma (202466_at), score: -0.56 POMT1protein-O-mannosyltransferase 1 (218476_at), score: -0.62 PPCDCphosphopantothenoylcysteine decarboxylase (219066_at), score: -0.45 PPP2R5Aprotein phosphatase 2, regulatory subunit B', alpha isoform (202187_s_at), score: -0.52 PPP2R5Dprotein phosphatase 2, regulatory subunit B', delta isoform (211159_s_at), score: 0.63 PPP5Cprotein phosphatase 5, catalytic subunit (201979_s_at), score: 0.42 PRKAB1protein kinase, AMP-activated, beta 1 non-catalytic subunit (201835_s_at), score: 0.62 PRPF6PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) (208879_x_at), score: 0.6 PRR4proline rich 4 (lacrimal) (204919_at), score: 0.55 PRRX1paired related homeobox 1 (205991_s_at), score: 0.56 PSENENpresenilin enhancer 2 homolog (C. elegans) (218302_at), score: 0.44 PSME4proteasome (prosome, macropain) activator subunit 4 (212220_at), score: 0.4 PTBP1polypyrimidine tract binding protein 1 (212016_s_at), score: 0.63 PTCD1pentatricopeptide repeat domain 1 (218956_s_at), score: -0.45 PTMSparathymosin (218045_x_at), score: 0.42 PTPN14protein tyrosine phosphatase, non-receptor type 14 (205503_at), score: 0.62 PTPRAprotein tyrosine phosphatase, receptor type, A (213799_s_at), score: 0.52 PXNpaxillin (211823_s_at), score: 0.71 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.42 RAB35RAB35, member RAS oncogene family (205461_at), score: 0.71 RAB5ARAB5A, member RAS oncogene family (206113_s_at), score: 0.51 RALGPS2Ral GEF with PH domain and SH3 binding motif 2 (220338_at), score: 0.4 RAPGEF6Rap guanine nucleotide exchange factor (GEF) 6 (219112_at), score: -0.61 RASA3RAS p21 protein activator 3 (206220_s_at), score: 0.49 RBBP9retinoblastoma binding protein 9 (221440_s_at), score: 0.54 RBM47RNA binding motif protein 47 (218035_s_at), score: -0.51 RCAN1regulator of calcineurin 1 (215253_s_at), score: 0.44 RECQL5RecQ protein-like 5 (34063_at), score: 0.44 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: 0.57 RFX7regulatory factor X, 7 (218430_s_at), score: 0.4 RHOBTB1Rho-related BTB domain containing 1 (212651_at), score: -0.44 RNF44ring finger protein 44 (203286_at), score: -0.52 RPS20ribosomal protein S20 (216247_at), score: 0.5 RRN3RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) (216902_s_at), score: 0.61 RSAD1radical S-adenosyl methionine domain containing 1 (218307_at), score: -0.56 RXRBretinoid X receptor, beta (215099_s_at), score: 0.65 SCARB2scavenger receptor class B, member 2 (201647_s_at), score: 0.46 SEMA4Gsema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G (219194_at), score: -0.44 SETD8SET domain containing (lysine methyltransferase) 8 (220200_s_at), score: 0.4 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.46 SHC1SHC (Src homology 2 domain containing) transforming protein 1 (201469_s_at), score: 0.43 SIM2single-minded homolog 2 (Drosophila) (206558_at), score: 0.4 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: -0.44 SIRT1sirtuin (silent mating type information regulation 2 homolog) 1 (S. cerevisiae) (218878_s_at), score: -0.44 SKIv-ski sarcoma viral oncogene homolog (avian) (204270_at), score: 0.43 SLC12A4solute carrier family 12 (potassium/chloride transporters), member 4 (209401_s_at), score: 0.74 SLC14A1solute carrier family 14 (urea transporter), member 1 (Kidd blood group) (205856_at), score: -0.49 SLC17A5solute carrier family 17 (anion/sugar transporter), member 5 (221041_s_at), score: 0.39 SLC25A21solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21 (220474_at), score: -0.44 SLC37A4solute carrier family 37 (glucose-6-phosphate transporter), member 4 (217289_s_at), score: 0.5 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (207043_s_at), score: 0.88 SOCS1suppressor of cytokine signaling 1 (210001_s_at), score: 0.58 SPPL2Bsignal peptide peptidase-like 2B (215833_s_at), score: 0.62 SRPRsignal recognition particle receptor (docking protein) (200917_s_at), score: 0.64 SSTR1somatostatin receptor 1 (208482_at), score: 0.47 ST6GALNAC4ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 (221551_x_at), score: 0.49 STAT2signal transducer and activator of transcription 2, 113kDa (205170_at), score: 0.46 STIP1stress-induced-phosphoprotein 1 (212009_s_at), score: 0.47 TAC3tachykinin 3 (219992_at), score: 0.47 TAF4TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa (213090_s_at), score: -0.71 TBL3transducin (beta)-like 3 (209820_s_at), score: -0.48 TBX21T-box 21 (220684_at), score: 0.48 TGFBR1transforming growth factor, beta receptor 1 (206943_at), score: 0.4 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.72 TNFRSF10Ctumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain (206222_at), score: 0.51 TNFRSF10Dtumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain (210654_at), score: 0.47 TNFRSF12Atumor necrosis factor receptor superfamily, member 12A (218368_s_at), score: 0.44 TNFRSF25tumor necrosis factor receptor superfamily, member 25 (219423_x_at), score: 0.52 TOM1L2target of myb1-like 2 (chicken) (214840_at), score: 0.46 TOP3Atopoisomerase (DNA) III alpha (214300_s_at), score: 0.47 TRIM25tripartite motif-containing 25 (206911_at), score: 0.59 TRMT2BTRM2 tRNA methyltransferase 2 homolog B (S. cerevisiae) (205238_at), score: 0.45 UPF3BUPF3 regulator of nonsense transcripts homolog B (yeast) (218757_s_at), score: -0.43 UTRNutrophin (213022_s_at), score: 0.5 VEGFAvascular endothelial growth factor A (211527_x_at), score: 0.49 WACWW domain containing adaptor with coiled-coil (219679_s_at), score: 0.61 WDR68WD repeat domain 68 (221745_at), score: 0.41 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: -0.55 ZFP36L2zinc finger protein 36, C3H type-like 2 (201367_s_at), score: 0.54 ZFP37zinc finger protein 37 homolog (mouse) (207068_at), score: 0.45 ZNF137zinc finger protein 137 (207394_at), score: -0.45 ZNF236zinc finger protein 236 (47571_at), score: -0.46 ZNF26zinc finger protein 26 (219595_at), score: -0.51 ZNF273zinc finger protein 273 (215239_x_at), score: 0.49 ZNF318zinc finger protein 318 (203521_s_at), score: -0.43 ZNF574zinc finger protein 574 (218762_at), score: -0.72
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |