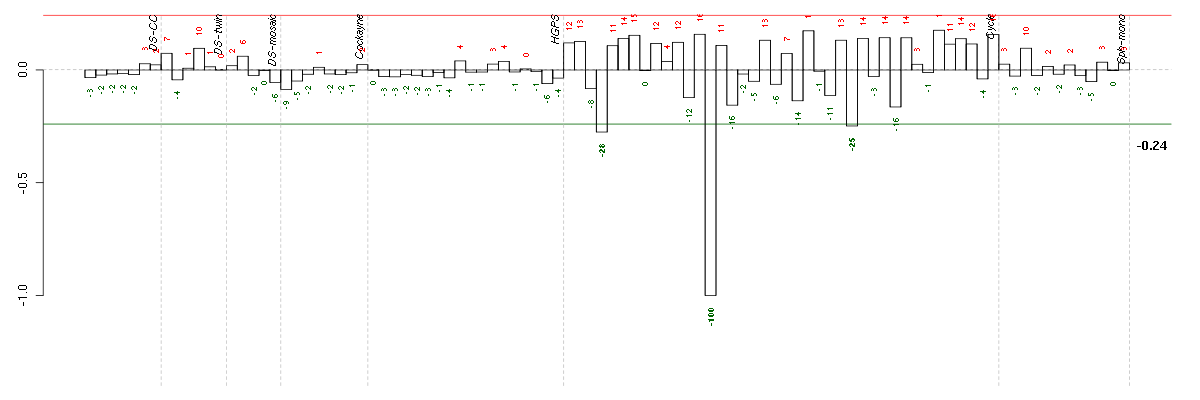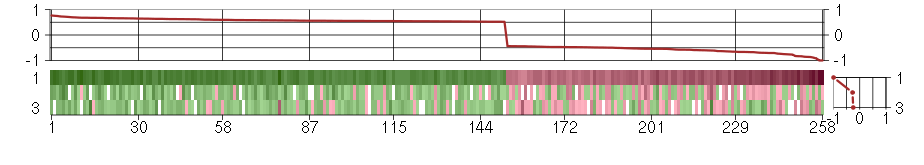

Under-expression is coded with green,
over-expression with red color.



ABI1abl-interactor 1 (209028_s_at), score: 0.55 ABT1activator of basal transcription 1 (218405_at), score: 0.58 ABTB2ankyrin repeat and BTB (POZ) domain containing 2 (213497_at), score: 0.7 ACSL6acyl-CoA synthetase long-chain family member 6 (211207_s_at), score: -0.68 ACTN3actinin, alpha 3 (206891_at), score: 0.67 ADAMDEC1ADAM-like, decysin 1 (206134_at), score: -0.85 ADAMTS9ADAM metallopeptidase with thrombospondin type 1 motif, 9 (220287_at), score: -0.63 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.54 AGAP2ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 (215080_s_at), score: -0.58 ALDH4A1aldehyde dehydrogenase 4 family, member A1 (211552_s_at), score: 0.58 ANGEL1angel homolog 1 (Drosophila) (36865_at), score: 0.56 ANKS1Aankyrin repeat and sterile alpha motif domain containing 1A (212747_at), score: 0.59 APBA1amyloid beta (A4) precursor protein-binding, family A, member 1 (206679_at), score: 0.54 APOC1apolipoprotein C-I (213553_x_at), score: 0.64 APPL1adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 (218158_s_at), score: 0.64 ASTN2astrotactin 2 (209693_at), score: 0.63 ATHL1ATH1, acid trehalase-like 1 (yeast) (219359_at), score: -0.6 ATMataxia telangiectasia mutated (210858_x_at), score: 0.56 ATP1B4ATPase, (Na+)/K+ transporting, beta 4 polypeptide (220556_at), score: -0.5 ATP8A1ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 (213106_at), score: -0.45 ATP8B2ATPase, class I, type 8B, member 2 (216873_s_at), score: 0.55 ATXN7ataxin 7 (204516_at), score: 0.59 AVILadvillin (205539_at), score: 0.75 BBS10Bardet-Biedl syndrome 10 (219487_at), score: 0.64 C10orf118chromosome 10 open reading frame 118 (219844_at), score: 0.54 C14orf135chromosome 14 open reading frame 135 (219972_s_at), score: 0.54 C1QTNF1C1q and tumor necrosis factor related protein 1 (220975_s_at), score: 0.61 C20orf111chromosome 20 open reading frame 111 (209020_at), score: 0.53 C20orf12chromosome 20 open reading frame 12 (219951_s_at), score: 0.61 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: 0.55 C3AR1complement component 3a receptor 1 (209906_at), score: 0.68 C4orf16chromosome 4 open reading frame 16 (219023_at), score: 0.55 C4orf29chromosome 4 open reading frame 29 (219980_at), score: -0.7 C5orf44chromosome 5 open reading frame 44 (218674_at), score: 0.58 C8orf44chromosome 8 open reading frame 44 (220216_at), score: 0.58 CCDC121coiled-coil domain containing 121 (220321_s_at), score: -0.54 CD200CD200 molecule (209583_s_at), score: -0.7 CD28CD28 molecule (206545_at), score: -0.54 CD53CD53 molecule (203416_at), score: 0.53 CDH19cadherin 19, type 2 (206898_at), score: -1 CHKAcholine kinase alpha (204233_s_at), score: 0.52 CHN2chimerin (chimaerin) 2 (207486_x_at), score: -0.66 CHRNA5cholinergic receptor, nicotinic, alpha 5 (206533_at), score: -0.51 CLTCL1clathrin, heavy chain-like 1 (205944_s_at), score: 0.68 CNNM1cyclin M1 (220166_at), score: 0.59 CNR2cannabinoid receptor 2 (macrophage) (206586_at), score: 0.55 COL9A2collagen, type IX, alpha 2 (213622_at), score: 0.56 COPS7BCOP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis) (219997_s_at), score: 0.52 COQ7coenzyme Q7 homolog, ubiquinone (yeast) (209745_at), score: -0.5 COX6A2cytochrome c oxidase subunit VIa polypeptide 2 (206353_at), score: 0.63 CPA4carboxypeptidase A4 (205832_at), score: 0.56 CPT2carnitine palmitoyltransferase 2 (204264_at), score: 0.53 CSTF2Tcleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant (212905_at), score: -0.49 CTSScathepsin S (202901_x_at), score: -0.92 CXCL11chemokine (C-X-C motif) ligand 11 (210163_at), score: -0.64 CYP26B1cytochrome P450, family 26, subfamily B, polypeptide 1 (219825_at), score: -0.67 CYSLTR1cysteinyl leukotriene receptor 1 (216288_at), score: -0.47 DCUN1D2DCN1, defective in cullin neddylation 1, domain containing 2 (S. cerevisiae) (219116_s_at), score: 0.57 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: 0.58 DNAL4dynein, axonemal, light chain 4 (204008_at), score: 0.53 DSCR6Down syndrome critical region gene 6 (207267_s_at), score: -0.83 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.63 DSTYKdual serine/threonine and tyrosine protein kinase (214663_at), score: 0.54 DTNAdystrobrevin, alpha (205741_s_at), score: -0.45 DTX2deltex homolog 2 (Drosophila) (215732_s_at), score: -0.48 DUSP8dual specificity phosphatase 8 (206374_at), score: 0.54 DYNC2H1dynein, cytoplasmic 2, heavy chain 1 (219469_at), score: 0.53 EIF2AK2eukaryotic translation initiation factor 2-alpha kinase 2 (204211_x_at), score: 0.56 EIF2C4eukaryotic translation initiation factor 2C, 4 (219190_s_at), score: 0.54 ELA3Aelastase 3A, pancreatic (211738_x_at), score: 0.54 ELLelongation factor RNA polymerase II (204095_s_at), score: 0.66 EML2echinoderm microtubule associated protein like 2 (204398_s_at), score: 0.59 EPHA3EPH receptor A3 (211164_at), score: -0.86 EPM2Aepilepsy, progressive myoclonus type 2A, Lafora disease (laforin) (205231_s_at), score: 0.6 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (214053_at), score: -0.73 ERN1endoplasmic reticulum to nucleus signaling 1 (207061_at), score: -0.46 EXD3exonuclease 3'-5' domain containing 3 (220838_at), score: 0.54 F7coagulation factor VII (serum prothrombin conversion accelerator) (207300_s_at), score: 0.57 FA2Hfatty acid 2-hydroxylase (219429_at), score: -0.44 FAM169Afamily with sequence similarity 169, member A (213954_at), score: -0.47 FBRSfibrosin (218255_s_at), score: 0.62 FBXO2F-box protein 2 (219305_x_at), score: 0.73 FBXO22F-box protein 22 (219638_at), score: -0.76 FCER1AFc fragment of IgE, high affinity I, receptor for; alpha polypeptide (211734_s_at), score: -0.53 FCGR2AFc fragment of IgG, low affinity IIa, receptor (CD32) (203561_at), score: -0.56 FGL1fibrinogen-like 1 (205305_at), score: 0.57 FKRPfukutin related protein (219853_at), score: -0.57 FNBP1Lformin binding protein 1-like (215017_s_at), score: 0.53 FOXK2forkhead box K2 (203064_s_at), score: -0.47 FRMD4BFERM domain containing 4B (213056_at), score: -0.75 GEMIN8gem (nuclear organelle) associated protein 8 (219252_s_at), score: -0.46 GLMNglomulin, FKBP associated protein (207153_s_at), score: 0.56 GLRA1glycine receptor, alpha 1 (207972_at), score: -0.57 GNL3LPguanine nucleotide binding protein-like 3 (nucleolar)-like pseudogene (220716_at), score: 0.63 GPR144G protein-coupled receptor 144 (216289_at), score: 0.58 GPR63G protein-coupled receptor 63 (220993_s_at), score: -0.44 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: 0.57 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: -0.64 GSTCDglutathione S-transferase, C-terminal domain containing (220063_at), score: -0.83 GSTZ1glutathione transferase zeta 1 (209531_at), score: 0.54 GTF2H3general transcription factor IIH, polypeptide 3, 34kDa (222104_x_at), score: 0.55 GYG2glycogenin 2 (215695_s_at), score: 0.57 HAB1B1 for mucin (215778_x_at), score: 0.62 HAMPhepcidin antimicrobial peptide (220491_at), score: 0.69 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: 0.72 HAVCR1hepatitis A virus cellular receptor 1 (207052_at), score: -0.47 HCG8HLA complex group 8 (215985_at), score: -1 HINFPhistone H4 transcription factor (206495_s_at), score: 0.52 HLA-DRB1major histocompatibility complex, class II, DR beta 1 (209312_x_at), score: -0.53 HOXA6homeobox A6 (208557_at), score: 0.59 HOXB6homeobox B6 (205366_s_at), score: -0.58 HRASLSHRAS-like suppressor (219983_at), score: -0.58 ICA1islet cell autoantigen 1, 69kDa (211740_at), score: -0.71 IGFALSinsulin-like growth factor binding protein, acid labile subunit (215712_s_at), score: 0.57 IKZF4IKAROS family zinc finger 4 (Eos) (208472_at), score: 0.62 INE1inactivation escape 1 (non-protein coding) (207252_at), score: 0.57 IRF6interferon regulatory factor 6 (202597_at), score: -0.49 ISOC1isochorismatase domain containing 1 (218170_at), score: 0.56 ITGA10integrin, alpha 10 (206766_at), score: 0.65 ITPKCinositol 1,4,5-trisphosphate 3-kinase C (213076_at), score: 0.52 JAK2Janus kinase 2 (a protein tyrosine kinase) (205842_s_at), score: -0.55 JMJD1Bjumonji domain containing 1B (210878_s_at), score: 0.55 JMJD1Cjumonji domain containing 1C (221763_at), score: 0.55 KCNIP1Kv channel interacting protein 1 (221307_at), score: -0.67 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: -0.54 KIAA0562KIAA0562 (204075_s_at), score: 0.56 KIAA0892KIAA0892 (212505_s_at), score: -0.46 KIAA1107KIAA1107 (214098_at), score: -0.6 KIF26Bkinesin family member 26B (220002_at), score: 0.67 KLF12Kruppel-like factor 12 (208467_at), score: -0.65 KLF7Kruppel-like factor 7 (ubiquitous) (204334_at), score: 0.56 KLHL3kelch-like 3 (Drosophila) (221221_s_at), score: -0.59 LAT2linker for activation of T cells family, member 2 (211768_at), score: 0.77 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: 0.66 LILRP2leukocyte immunoglobulin-like receptor pseudogene 2 (214561_at), score: -0.47 LIMD1LIM domains containing 1 (218850_s_at), score: -0.44 LOC100129500hypothetical protein LOC100129500 (212884_x_at), score: 0.53 LOC100130703similar to hCG2042168 (207596_at), score: -0.52 LOC100132247similar to Uncharacterized protein KIAA0220 (215002_at), score: -0.89 LOC149501similar to keratin 8 (216821_at), score: 0.65 LOC257152hypothetical protein LOC257152 (215302_at), score: -0.5 LOC729806similar to hCG1725380 (217544_at), score: -0.74 LOC93432maltase-glucoamylase-like pseudogene (216666_at), score: -0.65 MANBAmannosidase, beta A, lysosomal (203778_at), score: 0.75 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: -0.44 MAP3K9mitogen-activated protein kinase kinase kinase 9 (213927_at), score: -0.77 MAPRE3microtubule-associated protein, RP/EB family, member 3 (214270_s_at), score: 0.52 MCM3APASMCM3AP antisense RNA (non-protein coding) (220459_at), score: -0.66 MFSD6major facilitator superfamily domain containing 6 (219858_s_at), score: -0.49 MGC5590hypothetical protein MGC5590 (220931_at), score: -0.61 MID2midline 2 (208384_s_at), score: 0.54 MMP9matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (203936_s_at), score: 0.58 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: -0.48 MTMR3myotubularin related protein 3 (202197_at), score: -0.44 MUC3Amucin 3A, cell surface associated (217117_x_at), score: 0.63 MVKmevalonate kinase (36907_at), score: 0.53 MYBPC1myosin binding protein C, slow type (214087_s_at), score: -0.54 MYO1Amyosin IA (211916_s_at), score: -0.48 MYO5Cmyosin VC (218966_at), score: -0.71 MYRIPmyosin VIIA and Rab interacting protein (214156_at), score: -0.5 NAT9N-acetyltransferase 9 (GCN5-related, putative) (204382_at), score: 0.53 NCOA1nuclear receptor coactivator 1 (209106_at), score: 0.63 NEURLneuralized homolog (Drosophila) (204889_s_at), score: 0.53 NFRKBnuclear factor related to kappaB binding protein (213028_at), score: 0.68 NFYCnuclear transcription factor Y, gamma (202215_s_at), score: -0.48 NMUneuromedin U (206023_at), score: -0.63 NPIPL2nuclear pore complex interacting protein-like 2 (221992_at), score: 0.59 NSBP1nucleosomal binding protein 1 (221606_s_at), score: -0.51 OCEL1occludin/ELL domain containing 1 (205441_at), score: 0.54 OPLAH5-oxoprolinase (ATP-hydrolysing) (222025_s_at), score: 0.55 OR10C1olfactory receptor, family 10, subfamily C, member 1 (221339_at), score: -0.48 ORC1Lorigin recognition complex, subunit 1-like (yeast) (205085_at), score: -0.45 PAN2PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (203117_s_at), score: 0.54 PARD6Apar-6 partitioning defective 6 homolog alpha (C. elegans) (205245_at), score: 0.67 PAX8paired box 8 (121_at), score: 0.55 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: 0.55 PDE3Aphosphodiesterase 3A, cGMP-inhibited (206388_at), score: -0.86 PDZRN4PDZ domain containing ring finger 4 (220595_at), score: -0.44 PEX5peroxisomal biogenesis factor 5 (203244_at), score: 0.58 PGCprogastricsin (pepsinogen C) (205261_at), score: 0.56 PGPEP1pyroglutamyl-peptidase I (219891_at), score: 0.65 PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.59 PLEKHA9pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 9 (220157_x_at), score: 0.67 PLS1plastin 1 (I isoform) (205190_at), score: -0.67 PLXNB3plexin B3 (205957_at), score: -0.47 PMS2CLPMS2 C-terminal like pseudogene (221206_at), score: 0.56 PPIAL4Apeptidylprolyl isomerase A (cyclophilin A)-like 4A (217136_at), score: -0.66 PRKAB2protein kinase, AMP-activated, beta 2 non-catalytic subunit (214474_at), score: 0.62 PSD4pleckstrin and Sec7 domain containing 4 (203317_at), score: 0.66 R3HDM2R3H domain containing 2 (203831_at), score: 0.68 RAB26RAB26, member RAS oncogene family (50965_at), score: 0.6 RASIP1Ras interacting protein 1 (220027_s_at), score: 0.6 RBM12RNA binding motif protein 12 (212170_at), score: 0.58 RBM15RNA binding motif protein 15 (219286_s_at), score: 0.52 RECQL5RecQ protein-like 5 (34063_at), score: -0.66 RIN1Ras and Rab interactor 1 (205211_s_at), score: -0.44 RNASE2ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) (206111_at), score: -0.53 RP11-35N6.1plasticity related gene 3 (219732_at), score: -0.59 RPIAribose 5-phosphate isomerase A (212973_at), score: 0.62 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (220738_s_at), score: -0.54 RUFY3RUN and FYVE domain containing 3 (210251_s_at), score: 0.55 SAA4serum amyloid A4, constitutive (207096_at), score: 0.56 SAMD14sterile alpha motif domain containing 14 (213866_at), score: 0.52 SCARF1scavenger receptor class F, member 1 (206995_x_at), score: -0.52 SCN1Bsodium channel, voltage-gated, type I, beta (205508_at), score: 0.66 SERGEFsecretion regulating guanine nucleotide exchange factor (220482_s_at), score: 0.62 SERPINA7serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 (206386_at), score: 0.65 SF4splicing factor 4 (215004_s_at), score: 0.69 SFNstratifin (33322_i_at), score: 0.55 SH2D3ASH2 domain containing 3A (222169_x_at), score: 0.6 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: 0.66 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: -0.49 SLC12A3solute carrier family 12 (sodium/chloride transporters), member 3 (215274_at), score: -0.61 SLC12A9solute carrier family 12 (potassium/chloride transporters), member 9 (220371_s_at), score: 0.55 SLC16A6solute carrier family 16, member 6 (monocarboxylic acid transporter 7) (207038_at), score: -0.54 SLC2A3solute carrier family 2 (facilitated glucose transporter), member 3 (202499_s_at), score: 0.56 SLC35C2solute carrier family 35, member C2 (219447_s_at), score: 0.55 SLC43A1solute carrier family 43, member 1 (204394_at), score: 0.59 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.52 SLC6A14solute carrier family 6 (amino acid transporter), member 14 (219795_at), score: -0.71 SLC9A3R1solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 (201349_at), score: 0.66 SMA5glucuronidase, beta pseudogene (215043_s_at), score: 0.56 SNORA21small nucleolar RNA, H/ACA box 21 (215224_at), score: 0.54 SPAG16sperm associated antigen 16 (219109_at), score: 0.55 SPAM1sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding) (216989_at), score: -0.48 SPATA2Lspermatogenesis associated 2-like (214965_at), score: 0.58 SPON1spondin 1, extracellular matrix protein (209436_at), score: -0.47 SPTA1spectrin, alpha, erythrocytic 1 (elliptocytosis 2) (206937_at), score: -0.83 SSH3slingshot homolog 3 (Drosophila) (51192_at), score: 0.53 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (212894_at), score: 0.67 SYCP2synaptonemal complex protein 2 (206546_at), score: -0.54 TAAR2trace amine associated receptor 2 (221394_at), score: -0.75 THOC6THO complex 6 homolog (Drosophila) (218848_at), score: 0.58 TLR1toll-like receptor 1 (210176_at), score: -0.7 TMEM40transmembrane protein 40 (219503_s_at), score: 0.53 TRAPPC9trafficking protein particle complex 9 (221836_s_at), score: 0.71 TRIM34tripartite motif-containing 34 (221044_s_at), score: -0.57 TRIM45tripartite motif-containing 45 (219923_at), score: 0.62 TROtrophinin (211700_s_at), score: 0.55 TSPAN9tetraspanin 9 (220968_s_at), score: 0.66 TSPY1testis specific protein, Y-linked 1 (216374_at), score: -0.48 TWISTNBTWIST neighbor (214729_at), score: -0.6 UBE2D1ubiquitin-conjugating enzyme E2D 1 (UBC4/5 homolog, yeast) (211764_s_at), score: 0.52 USP6NLUSP6 N-terminal like (204761_at), score: 0.56 WWC1WW and C2 domain containing 1 (213085_s_at), score: 0.6 ZBTB10zinc finger and BTB domain containing 10 (219312_s_at), score: 0.55 ZBTB40zinc finger and BTB domain containing 40 (203958_s_at), score: 0.64 ZCCHC4zinc finger, CCHC domain containing 4 (220473_s_at), score: -0.49 ZCCHC6zinc finger, CCHC domain containing 6 (220933_s_at), score: 0.54 ZKSCAN5zinc finger with KRAB and SCAN domains 5 (203730_s_at), score: -0.61 ZNF133zinc finger protein 133 (216960_s_at), score: 0.63 ZNF175zinc finger protein 175 (205497_at), score: -0.54 ZNF329zinc finger protein 329 (219765_at), score: 0.52 ZNF529zinc finger protein 529 (215307_at), score: 0.62 ZNF614zinc finger protein 614 (220721_at), score: -0.47 ZNF652zinc finger protein 652 (205594_at), score: 0.64 ZNF749zinc finger protein 749 (215289_at), score: -0.51 ZNF783zinc finger family member 783 (221876_at), score: 0.54
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |