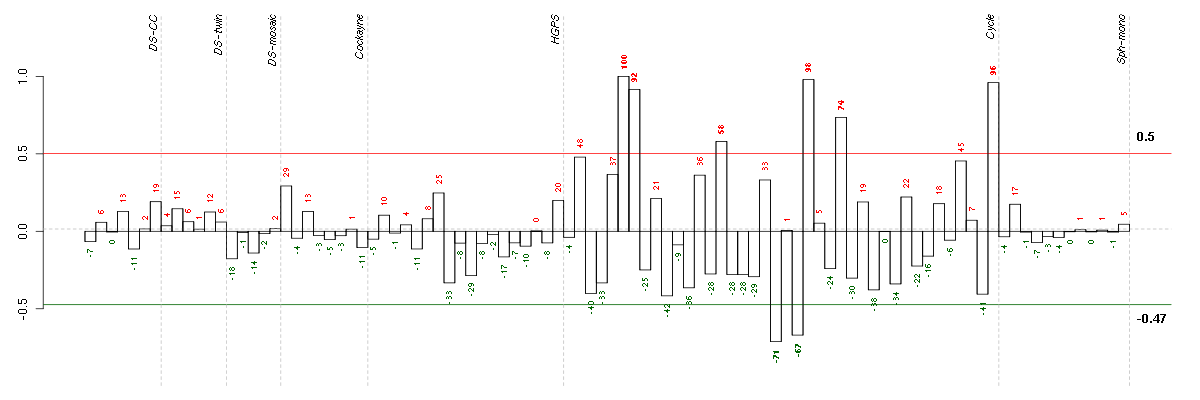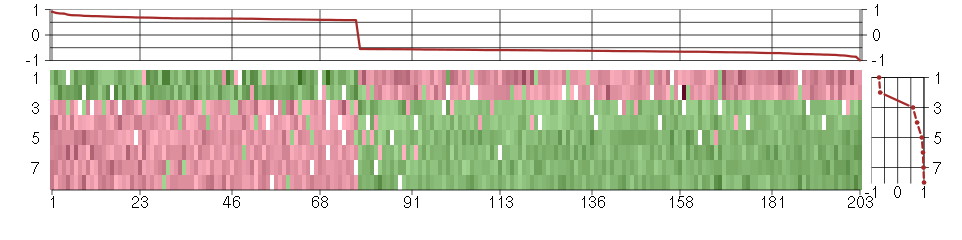

Under-expression is coded with green,
over-expression with red color.



ABI3BPABI family, member 3 (NESH) binding protein (220518_at), score: -0.55 ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.7 ACTN3actinin, alpha 3 (206891_at), score: 0.65 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.76 ADCY3adenylate cyclase 3 (209321_s_at), score: -0.82 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: -0.57 AKR1D1aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) (207102_at), score: -0.55 ANAPC2anaphase promoting complex subunit 2 (218555_at), score: -0.61 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (217888_s_at), score: -0.56 ARHGAP22Rho GTPase activating protein 22 (206298_at), score: -0.6 ARL17P1ADP-ribosylation factor-like 17 pseudogene 1 (210718_s_at), score: -0.68 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: -0.61 ATN1atrophin 1 (40489_at), score: -0.65 ATXN1ataxin 1 (203232_s_at), score: 0.64 BIN1bridging integrator 1 (210201_x_at), score: -0.58 BRCA2breast cancer 2, early onset (208368_s_at), score: 0.72 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.62 C10orf26chromosome 10 open reading frame 26 (202808_at), score: -0.59 C10orf81chromosome 10 open reading frame 81 (219857_at), score: -0.66 C14orf106chromosome 14 open reading frame 106 (206500_s_at), score: 0.64 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: -0.72 C19orf40chromosome 19 open reading frame 40 (214816_x_at), score: 0.7 C1orf38chromosome 1 open reading frame 38 (210785_s_at), score: -0.63 C5orf28chromosome 5 open reading frame 28 (219029_at), score: 0.58 CCDC101coiled-coil domain containing 101 (48117_at), score: 0.62 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.63 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.71 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: -0.6 CEP192centrosomal protein 192kDa (218827_s_at), score: 0.64 CEP57centrosomal protein 57kDa (203491_s_at), score: 0.63 CHRNA5cholinergic receptor, nicotinic, alpha 5 (206533_at), score: -0.56 CHST2carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 (203921_at), score: -0.74 CICcapicua homolog (Drosophila) (212784_at), score: -0.59 CLIP1CAP-GLY domain containing linker protein 1 (210716_s_at), score: 0.65 CLIP2CAP-GLY domain containing linker protein 2 (211031_s_at), score: -0.6 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: -0.58 CNNM2cyclin M2 (206818_s_at), score: 0.61 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.63 COL10A1collagen, type X, alpha 1 (217428_s_at), score: -0.77 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: -0.73 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -0.8 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.66 CUX1cut-like homeobox 1 (214743_at), score: 0.59 CYP7B1cytochrome P450, family 7, subfamily B, polypeptide 1 (207386_at), score: -0.69 CYTH4cytohesin 4 (219183_s_at), score: 0.66 DDX54DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 (219111_s_at), score: -0.57 DENND4BDENN/MADD domain containing 4B (202860_at), score: -0.77 DESdesmin (216947_at), score: -0.67 DKFZP586I1420hypothetical protein DKFZp586I1420 (213546_at), score: 0.7 DLSTPdihydrolipoamide S-succinyltransferase pseudogene (E2 component of 2-oxo-glutarate complex) (215210_s_at), score: 0.59 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: -0.74 DOK4docking protein 4 (209691_s_at), score: -0.55 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.61 EML2echinoderm microtubule associated protein like 2 (204398_s_at), score: 0.64 EPHA4EPH receptor A4 (206114_at), score: 0.61 F11coagulation factor XI (206610_s_at), score: 0.64 FAM3Afamily with sequence similarity 3, member A (38043_at), score: -0.56 FASNfatty acid synthase (212218_s_at), score: -0.65 FBXO17F-box protein 17 (220233_at), score: -0.58 FKBPLFK506 binding protein like (219187_at), score: 0.59 FKRPfukutin related protein (219853_at), score: -0.6 FKSG2apoptosis inhibitor (208588_at), score: 0.62 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.66 FLJ20674hypothetical protein FLJ20674 (220137_at), score: 0.64 FLJ40113golgi autoantigen, golgin subfamily a-like pseudogene (213212_x_at), score: 0.64 FOXK2forkhead box K2 (203064_s_at), score: -1 GATCglutamyl-tRNA(Gln) amidotransferase, subunit C homolog (bacterial) (214711_at), score: 0.59 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: -0.56 GGTLC1gamma-glutamyltransferase light chain 1 (211416_x_at), score: 0.67 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: -0.85 GP1BAglycoprotein Ib (platelet), alpha polypeptide (207389_at), score: 0.74 GP1BBglycoprotein Ib (platelet), beta polypeptide (206655_s_at), score: -0.78 HAB1B1 for mucin (215778_x_at), score: 0.65 HECW1HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 (215584_at), score: 0.68 HIST1H2BHhistone cluster 1, H2bh (208546_x_at), score: 0.63 HLA-DMBmajor histocompatibility complex, class II, DM beta (203932_at), score: 0.58 HNRNPA3P1heterogeneous nuclear ribonucleoprotein A3 pseudogene 1 (206809_s_at), score: -0.61 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.73 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: -0.62 HTR2B5-hydroxytryptamine (serotonin) receptor 2B (206638_at), score: -0.56 IDUAiduronidase, alpha-L- (205059_s_at), score: -0.75 IL9Rinterleukin 9 receptor (217212_s_at), score: 0.65 KATNB1katanin p80 (WD repeat containing) subunit B 1 (203162_s_at), score: 0.65 KCNN2potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 (220116_at), score: 0.84 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: -0.62 KIAA0317KIAA0317 (202128_at), score: -0.7 KIAA1009KIAA1009 (206005_s_at), score: 0.87 KIAA1305KIAA1305 (220911_s_at), score: -0.69 KRT24keratin 24 (220267_at), score: 0.75 LARSleucyl-tRNA synthetase (217810_x_at), score: 0.59 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: 0.66 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: 0.77 LOC128192similar to peptidyl-Pro cis trans isomerase (217346_at), score: 0.74 LOC80054hypothetical LOC80054 (220465_at), score: 0.62 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: -0.72 LRRC37A2leucine rich repeat containing 37, member A2 (221740_x_at), score: 0.71 MAFKv-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian) (206750_at), score: 0.64 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.57 MAP2K3mitogen-activated protein kinase kinase 3 (207667_s_at), score: -0.63 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: -0.68 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: -0.85 MC4Rmelanocortin 4 receptor (221467_at), score: 0.68 MEN1multiple endocrine neoplasia I (202645_s_at), score: -0.56 MICAL3microtubule associated monoxygenase, calponin and LIM domain containing 3 (212715_s_at), score: -0.68 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: -0.64 MLL4myeloid/lymphoid or mixed-lineage leukemia 4 (203419_at), score: -0.61 MOAP1modulator of apoptosis 1 (212508_at), score: 0.64 MR1major histocompatibility complex, class I-related (207565_s_at), score: 0.6 MUC3Bmucin 3B, cell surface associated (214898_x_at), score: 0.59 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: -0.65 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: 0.65 MYST3MYST histone acetyltransferase (monocytic leukemia) 3 (202423_at), score: -0.57 NAALAD2N-acetylated alpha-linked acidic dipeptidase 2 (222161_at), score: -0.6 NCKAP1NCK-associated protein 1 (207738_s_at), score: 0.67 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: -0.63 NFYCnuclear transcription factor Y, gamma (202215_s_at), score: -0.59 NUDT11nudix (nucleoside diphosphate linked moiety X)-type motif 11 (219855_at), score: -0.65 NUPL2nucleoporin like 2 (204003_s_at), score: 0.69 OPRL1opiate receptor-like 1 (206564_at), score: 0.64 OR7E156Polfactory receptor, family 7, subfamily E, member 156 pseudogene (222327_x_at), score: 0.65 ORAI2ORAI calcium release-activated calcium modulator 2 (218812_s_at), score: -0.58 PARVBparvin, beta (37965_at), score: -0.62 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: -0.62 PCF11PCF11, cleavage and polyadenylation factor subunit, homolog (S. cerevisiae) (203378_at), score: -0.55 PDLIM4PDZ and LIM domain 4 (211564_s_at), score: -0.68 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: -0.57 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: -0.62 PISDphosphatidylserine decarboxylase (202392_s_at), score: -0.64 PKN1protein kinase N1 (202161_at), score: -0.64 PLAGL2pleiomorphic adenoma gene-like 2 (202925_s_at), score: -0.68 PLCD1phospholipase C, delta 1 (205125_at), score: -0.56 PLCE1phospholipase C, epsilon 1 (205112_at), score: 0.65 POLHpolymerase (DNA directed), eta (219380_x_at), score: 0.78 POLRMTpolymerase (RNA) mitochondrial (DNA directed) (203782_s_at), score: -0.59 POLSpolymerase (DNA directed) sigma (202466_at), score: -0.58 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.66 POP1processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) (213449_at), score: -0.56 PRKCDprotein kinase C, delta (202545_at), score: -0.68 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: -0.55 PRR14proline rich 14 (218714_at), score: -0.59 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: -0.66 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: -0.64 RAB11FIP3RAB11 family interacting protein 3 (class II) (203933_at), score: -0.57 RAB22ARAB22A, member RAS oncogene family (218360_at), score: -0.68 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: -0.8 RBBP9retinoblastoma binding protein 9 (221440_s_at), score: 0.61 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: -0.61 RNF128ring finger protein 128 (219263_at), score: -0.6 RNF187ring finger protein 187 (212155_at), score: -0.61 RNF220ring finger protein 220 (219988_s_at), score: -0.63 RNF6ring finger protein (C3H2C3 type) 6 (210932_s_at), score: 0.6 RPL15P22ribosomal protein L15 pseudogene 22 (217266_at), score: 0.6 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.84 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.77 RSAD1radical S-adenosyl methionine domain containing 1 (218307_at), score: -0.59 S100A14S100 calcium binding protein A14 (218677_at), score: 0.61 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: -0.59 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.66 SF1splicing factor 1 (208313_s_at), score: -0.61 SHQ1SHQ1 homolog (S. cerevisiae) (63009_at), score: -0.75 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: -0.71 SIPA1L3signal-induced proliferation-associated 1 like 3 (213600_at), score: -0.62 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: -0.78 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: -0.71 SMYD5SMYD family member 5 (209516_at), score: -0.65 SNX27sorting nexin family member 27 (221006_s_at), score: -0.61 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.75 SPATA6spermatogenesis associated 6 (220298_s_at), score: 0.58 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: -0.57 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: -0.59 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: -0.59 ST7suppression of tumorigenicity 7 (207871_s_at), score: 0.72 STAU2staufen, RNA binding protein, homolog 2 (Drosophila) (204226_at), score: 0.65 TAOK2TAO kinase 2 (204986_s_at), score: -0.62 TAPBPTAP binding protein (tapasin) (208829_at), score: -0.66 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: -0.58 TBX3T-box 3 (219682_s_at), score: -0.64 TCEB3Btranscription elongation factor B polypeptide 3B (elongin A2) (220844_at), score: -0.6 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.58 TLE6transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) (222219_s_at), score: 0.65 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: 0.66 TMEM149transmembrane protein 149 (219690_at), score: 0.92 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.71 TNIP1TNFAIP3 interacting protein 1 (207196_s_at), score: -0.65 TOP3Atopoisomerase (DNA) III alpha (214300_s_at), score: 0.75 TSKUtsukushin (218245_at), score: -0.67 TXLNAtaxilin alpha (212300_at), score: -0.59 UBTD1ubiquitin domain containing 1 (219172_at), score: -0.67 UNC84Aunc-84 homolog A (C. elegans) (212074_at), score: 0.62 USP20ubiquitin specific peptidase 20 (203965_at), score: -0.56 UTRNutrophin (213022_s_at), score: 0.68 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: -0.76 WDR42AWD repeat domain 42A (202249_s_at), score: -0.57 WDR6WD repeat domain 6 (217734_s_at), score: -0.63 WFDC2WAP four-disulfide core domain 2 (203892_at), score: 0.6 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: 0.69 XAB2XPA binding protein 2 (218110_at), score: -0.71 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: 0.74 ZDHHC14zinc finger, DHHC-type containing 14 (219247_s_at), score: 0.79 ZNF318zinc finger protein 318 (203521_s_at), score: -0.56 ZNF529zinc finger protein 529 (215307_at), score: 0.71 ZNF580zinc finger protein 580 (220748_s_at), score: -0.64 ZNF702Pzinc finger protein 702 pseudogene (206557_at), score: -0.55
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |