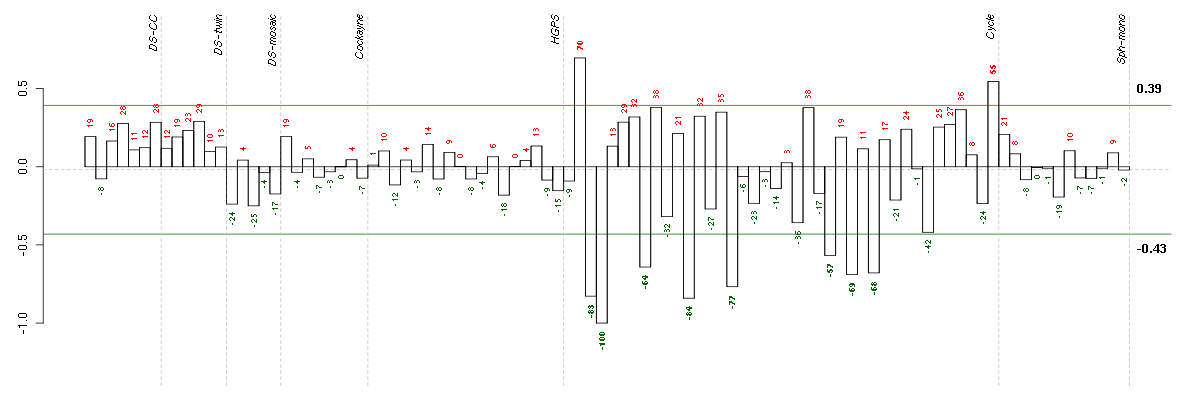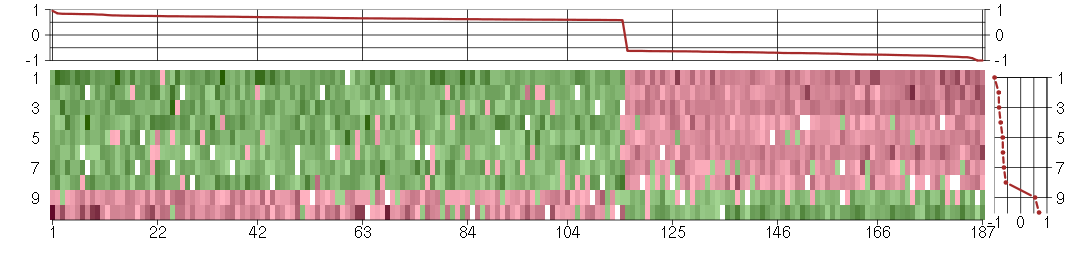

Under-expression is coded with green,
over-expression with red color.

biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
positive regulation of catalytic activity
Any process that activates or increases the activity of an enzyme.
regulation of MAP kinase activity
Any process that modulates the frequency, rate or extent of MAP kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
regulation of JUN kinase activity
Any process that modulates the frequency, rate or extent of JUN kinase activity.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.
regulation of kinase activity
Any process that modulates the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
regulation of protein kinase activity
Any process that modulates the frequency, rate or extent of protein kinase activity.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of transferase activity
Any process that modulates the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of molecular functions. Molecular functions are elemental biological activities occurring at the molecular level, such as catalysis or binding.
all
This term is the most general term possible
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.

ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: 0.95 ABCC3ATP-binding cassette, sub-family C (CFTR/MRP), member 3 (208161_s_at), score: 0.64 ABCC9ATP-binding cassette, sub-family C (CFTR/MRP), member 9 (208561_at), score: -0.77 ABCG4ATP-binding cassette, sub-family G (WHITE), member 4 (207593_at), score: 0.69 ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.68 ACTN3actinin, alpha 3 (206891_at), score: 0.66 ADAadenosine deaminase (204639_at), score: 0.66 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: 0.65 ADORA1adenosine A1 receptor (216220_s_at), score: 0.7 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: 0.73 ALDH3A1aldehyde dehydrogenase 3 family, memberA1 (205623_at), score: 0.74 ALPPL2alkaline phosphatase, placental-like 2 (216377_x_at), score: 0.72 ANXA8L2annexin A8-like 2 (203074_at), score: 0.6 AP1G2adaptor-related protein complex 1, gamma 2 subunit (201613_s_at), score: 0.59 ARHGDIBRho GDP dissociation inhibitor (GDI) beta (201288_at), score: -0.66 ARL8BADP-ribosylation factor-like 8B (217852_s_at), score: 0.66 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: 0.65 ATHL1ATH1, acid trehalase-like 1 (yeast) (219359_at), score: 0.59 ATMataxia telangiectasia mutated (210858_x_at), score: 0.72 BAT2HLA-B associated transcript 2 (212081_x_at), score: -0.78 BCL2L1BCL2-like 1 (215037_s_at), score: -0.72 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: -0.63 C2orf37chromosome 2 open reading frame 37 (220172_at), score: -0.87 C7orf58chromosome 7 open reading frame 58 (220032_at), score: -0.87 CALB2calbindin 2 (205428_s_at), score: 0.78 CAPN5calpain 5 (205166_at), score: -0.64 CATSPER2cation channel, sperm associated 2 (217588_at), score: 0.63 CCDC40coiled-coil domain containing 40 (220592_at), score: 0.69 CCDC94coiled-coil domain containing 94 (204335_at), score: 0.61 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.82 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.7 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: -0.73 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: -0.73 CMAHcytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) pseudogene (205518_s_at), score: -0.71 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.75 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.61 COL16A1collagen, type XVI, alpha 1 (204345_at), score: 0.69 CPNE3copine III (202118_s_at), score: -0.73 CTNScystinosis, nephropathic (36566_at), score: 0.59 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: 0.73 DCBLD2discoidin, CUB and LCCL domain containing 2 (213865_at), score: -0.66 DENND3DENN/MADD domain containing 3 (212974_at), score: -0.75 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: 0.62 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: 0.75 DNAJC2DnaJ (Hsp40) homolog, subfamily C, member 2 (213097_s_at), score: 0.69 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.64 EMCNendomucin (219436_s_at), score: -0.8 EPHA4EPH receptor A4 (206114_at), score: 0.64 EPM2Aepilepsy, progressive myoclonus type 2A, Lafora disease (laforin) (205231_s_at), score: 0.73 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (207347_at), score: -0.65 EVLEnah/Vasp-like (217838_s_at), score: -0.68 F11coagulation factor XI (206610_s_at), score: 0.65 FAM105Afamily with sequence similarity 105, member A (219694_at), score: -0.63 FAT1FAT tumor suppressor homolog 1 (Drosophila) (201579_at), score: 0.62 FBP1fructose-1,6-bisphosphatase 1 (209696_at), score: 0.72 FGL1fibrinogen-like 1 (205305_at), score: 0.65 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.82 FLJ21075hypothetical protein FLJ21075 (221172_at), score: 0.63 FLRT3fibronectin leucine rich transmembrane protein 3 (219250_s_at), score: -0.86 GABARAPL1GABA(A) receptor-associated protein like 1 (208868_s_at), score: 0.8 GABARAPL3GABA(A) receptors associated protein like 3 (pseudogene) (211458_s_at), score: 0.83 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: 0.83 GCHFRGTP cyclohydrolase I feedback regulator (204867_at), score: 0.7 GDF9growth differentiation factor 9 (221314_at), score: 0.59 GNL3Lguanine nucleotide binding protein-like 3 (nucleolar)-like (205010_at), score: 0.6 GOLGA8Ggolgi autoantigen, golgin subfamily a, 8G (222149_x_at), score: 0.69 GPR144G protein-coupled receptor 144 (216289_at), score: 0.82 GPR56G protein-coupled receptor 56 (212070_at), score: -0.81 HAB1B1 for mucin (215778_x_at), score: 0.82 HAMPhepcidin antimicrobial peptide (220491_at), score: 0.75 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: 0.62 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: 0.61 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.76 HIST1H1Chistone cluster 1, H1c (209398_at), score: -0.75 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.82 HSPA9heat shock 70kDa protein 9 (mortalin) (200690_at), score: 0.6 HUNKhormonally up-regulated Neu-associated kinase (219535_at), score: 0.6 IDI2isopentenyl-diphosphate delta isomerase 2 (217631_at), score: 0.68 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: 0.8 IGHMBP2immunoglobulin mu binding protein 2 (215980_s_at), score: 0.59 IL9Rinterleukin 9 receptor (217212_s_at), score: 0.77 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: -0.83 INE1inactivation escape 1 (non-protein coding) (207252_at), score: 0.62 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.63 JAG2jagged 2 (32137_at), score: 0.72 JUPjunction plakoglobin (201015_s_at), score: -0.79 KCTD15potassium channel tetramerisation domain containing 15 (218553_s_at), score: 0.73 KIF26Bkinesin family member 26B (220002_at), score: 0.74 KLF7Kruppel-like factor 7 (ubiquitous) (204334_at), score: 0.68 LEPREL2leprecan-like 2 (204854_at), score: -0.79 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: 0.72 LMO2LIM domain only 2 (rhombotin-like 1) (204249_s_at), score: -0.63 LOC100133748similar to GTF2IRD2 protein (215569_at), score: 0.63 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.71 LOC81691exonuclease NEF-sp (208107_s_at), score: -0.64 LPHN1latrophilin 1 (203488_at), score: 0.67 LRP2low density lipoprotein-related protein 2 (205710_at), score: -0.77 MAFGv-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) (204970_s_at), score: 0.61 MAP3K2mitogen-activated protein kinase kinase kinase 2 (221695_s_at), score: -0.66 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: -0.73 MEG3maternally expressed 3 (non-protein coding) (210794_s_at), score: 0.73 MEOX1mesenchyme homeobox 1 (205619_s_at), score: 0.85 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: 0.73 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: -0.71 MPP2membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) (213270_at), score: 0.59 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: -0.81 MTMR11myotubularin related protein 11 (205076_s_at), score: -0.71 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.64 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: 0.6 NAB2NGFI-A binding protein 2 (EGR1 binding protein 2) (216017_s_at), score: -0.7 NARG1LNMDA receptor regulated 1-like (219378_at), score: 0.6 NDPNorrie disease (pseudoglioma) (206022_at), score: -0.83 NF1neurofibromin 1 (211094_s_at), score: -0.78 NMT2N-myristoyltransferase 2 (205005_s_at), score: -0.62 NOL10nucleolar protein 10 (218591_s_at), score: 0.65 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: 0.73 NPY2Rneuropeptide Y receptor Y2 (210730_s_at), score: 0.58 NXPH3neurexophilin 3 (221991_at), score: 0.75 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: -0.77 OLFML2Bolfactomedin-like 2B (213125_at), score: -1 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: 0.75 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: -0.76 PARD6Apar-6 partitioning defective 6 homolog alpha (C. elegans) (205245_at), score: 0.63 PARP6poly (ADP-ribose) polymerase family, member 6 (219639_x_at), score: 0.58 PARVBparvin, beta (37965_at), score: -0.69 PCNXpecanex homolog (Drosophila) (213159_at), score: -0.63 PCSK5proprotein convertase subtilisin/kexin type 5 (205559_s_at), score: 0.61 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: -0.85 PIP4K2Cphosphatidylinositol-5-phosphate 4-kinase, type II, gamma (218942_at), score: 0.61 PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.6 PLAUplasminogen activator, urokinase (211668_s_at), score: -0.67 PMLpromyelocytic leukemia (206503_x_at), score: -0.68 PNMAL1PNMA-like 1 (218824_at), score: -0.76 PPP1R13Bprotein phosphatase 1, regulatory (inhibitor) subunit 13B (216347_s_at), score: 0.83 PRRX1paired related homeobox 1 (205991_s_at), score: -0.66 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.77 PXNpaxillin (211823_s_at), score: -0.91 RASGRP1RAS guanyl releasing protein 1 (calcium and DAG-regulated) (205590_at), score: 0.59 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: -1 RBM38RNA binding motif protein 38 (212430_at), score: 0.6 REG3Aregenerating islet-derived 3 alpha (205815_at), score: 0.68 RNF121ring finger protein 121 (219021_at), score: 0.61 RNF220ring finger protein 220 (219988_s_at), score: -0.63 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.71 RPS6KA4ribosomal protein S6 kinase, 90kDa, polypeptide 4 (204632_at), score: -0.67 RSAD1radical S-adenosyl methionine domain containing 1 (218307_at), score: -0.63 SAA4serum amyloid A4, constitutive (207096_at), score: 0.71 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: -0.67 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.71 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: -0.84 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: 0.71 SH2B3SH2B adaptor protein 3 (203320_at), score: -0.62 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: -0.65 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: 0.64 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: 0.59 SLC26A10solute carrier family 26, member 10 (214951_at), score: 0.75 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: -0.72 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: -0.64 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.76 SNTG1syntrophin, gamma 1 (220405_at), score: -0.64 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.69 SORBS1sorbin and SH3 domain containing 1 (218087_s_at), score: 0.68 SORL1sortilin-related receptor, L(DLR class) A repeats-containing (203509_at), score: 0.62 SPAG8sperm associated antigen 8 (206816_s_at), score: 0.65 SPATA1spermatogenesis associated 1 (221057_at), score: 0.7 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.66 SYT1synaptotagmin I (203999_at), score: -0.71 TAL1T-cell acute lymphocytic leukemia 1 (206283_s_at), score: 0.62 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: -0.81 TGM4transglutaminase 4 (prostate) (217566_s_at), score: 0.7 TMPRSS6transmembrane protease, serine 6 (214955_at), score: 0.66 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.72 TNK1tyrosine kinase, non-receptor, 1 (217149_x_at), score: 0.6 TOM1L2target of myb1-like 2 (chicken) (214840_at), score: -0.66 TRAPPC6Atrafficking protein particle complex 6A (204985_s_at), score: 0.63 TRIM34tripartite motif-containing 34 (221044_s_at), score: -0.64 TULP2tubby like protein 2 (206733_at), score: 0.7 UTF1undifferentiated embryonic cell transcription factor 1 (208275_x_at), score: 0.61 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: 0.6 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.79 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: 0.74 ZAKsterile alpha motif and leucine zipper containing kinase AZK (218833_at), score: 0.64 ZNF185zinc finger protein 185 (LIM domain) (203585_at), score: -0.68 ZNF281zinc finger protein 281 (218401_s_at), score: 0.61 ZNF510zinc finger protein 510 (206053_at), score: -0.67 ZNF783zinc finger family member 783 (221876_at), score: 0.67
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |