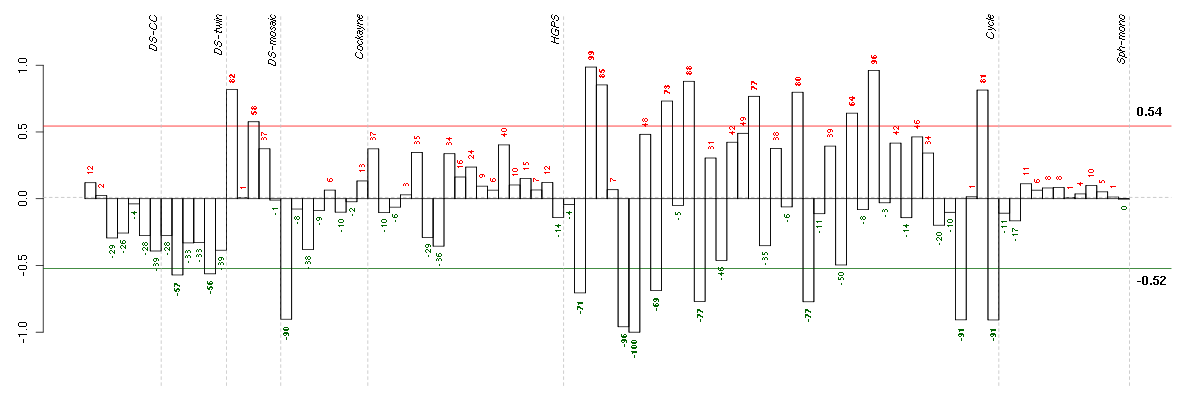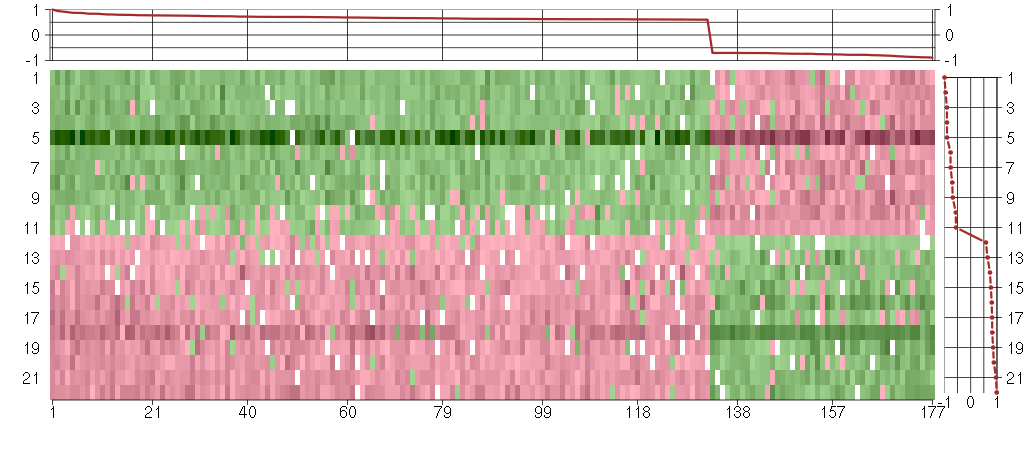

Under-expression is coded with green,
over-expression with red color.



ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.76 ACTN3actinin, alpha 3 (206891_at), score: -0.76 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.62 AGRNagrin (212285_s_at), score: 0.61 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.87 ARSAarylsulfatase A (204443_at), score: 0.77 ATF5activating transcription factor 5 (204999_s_at), score: 0.62 ATN1atrophin 1 (40489_at), score: 0.63 ATP6V0A1ATPase, H+ transporting, lysosomal V0 subunit a1 (205095_s_at), score: 0.61 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: 0.63 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.71 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.84 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.75 BCL2L1BCL2-like 1 (215037_s_at), score: 0.64 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.67 CALB2calbindin 2 (205428_s_at), score: -0.86 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.69 CCDC41coiled-coil domain containing 41 (219644_at), score: -0.71 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.74 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: 0.67 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: 0.64 CICcapicua homolog (Drosophila) (212784_at), score: 0.7 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.71 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.65 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.66 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: -0.8 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: 0.7 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.7 CUL7cullin 7 (36084_at), score: 0.6 DENND3DENN/MADD domain containing 3 (212974_at), score: 0.71 DGCR2DiGeorge syndrome critical region gene 2 (214198_s_at), score: -0.71 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.79 DNAJC4DnaJ (Hsp40) homolog, subfamily C, member 4 (206782_s_at), score: 0.61 DNM2dynamin 2 (202253_s_at), score: 0.62 DOK4docking protein 4 (209691_s_at), score: 0.65 DOPEY1dopey family member 1 (40612_at), score: -0.78 DRG2developmentally regulated GTP binding protein 2 (203267_s_at), score: 0.66 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.74 ECE1endothelin converting enzyme 1 (201750_s_at), score: 0.62 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.61 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: 0.62 EPHA4EPH receptor A4 (206114_at), score: -0.84 EVLEnah/Vasp-like (217838_s_at), score: 0.6 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.76 FBXO17F-box protein 17 (220233_at), score: 0.64 FBXO2F-box protein 2 (219305_x_at), score: -0.78 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: -0.78 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.7 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.79 FKBP8FK506 binding protein 8, 38kDa (208255_s_at), score: 0.62 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.77 FOXF1forkhead box F1 (205935_at), score: 0.65 FOXK2forkhead box K2 (203064_s_at), score: 0.9 GMPRguanosine monophosphate reductase (204187_at), score: 0.71 GTPBP1GTP binding protein 1 (219357_at), score: 0.64 HAB1B1 for mucin (215778_x_at), score: -0.89 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.77 HECTD3HECT domain containing 3 (218632_at), score: 0.61 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: -0.71 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: 0.62 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.73 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.78 HSF1heat shock transcription factor 1 (202344_at), score: 0.77 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.83 IL9Rinterleukin 9 receptor (217212_s_at), score: -0.83 INPP1inositol polyphosphate-1-phosphatase (202794_at), score: 0.62 JUNDjun D proto-oncogene (203751_x_at), score: 0.77 JUPjunction plakoglobin (201015_s_at), score: 0.87 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.82 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: 0.62 KIAA1305KIAA1305 (220911_s_at), score: 0.72 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.73 KLHL26kelch-like 26 (Drosophila) (219354_at), score: 0.68 LEPREL2leprecan-like 2 (204854_at), score: 0.82 LIMS2LIM and senescent cell antigen-like domains 2 (220765_s_at), score: 0.62 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.92 MAP2K2mitogen-activated protein kinase kinase 2 (213490_s_at), score: 0.62 MAP2K3mitogen-activated protein kinase kinase 3 (207667_s_at), score: 0.68 MAP3K11mitogen-activated protein kinase kinase kinase 11 (203652_at), score: 0.61 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.77 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: 0.63 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.71 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.74 MYO15Amyosin XVA (220288_at), score: -0.71 NACAP1nascent-polypeptide-associated complex alpha polypeptide pseudogene 1 (211445_x_at), score: -0.7 NCDNneurochondrin (209556_at), score: 0.63 NCKAP1NCK-associated protein 1 (207738_s_at), score: -0.72 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: -0.71 NEFMneurofilament, medium polypeptide (205113_at), score: -0.75 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.66 NINJ1ninjurin 1 (203045_at), score: 0.73 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.79 NPDC1neural proliferation, differentiation and control, 1 (218086_at), score: 0.67 NXPH3neurexophilin 3 (221991_at), score: -0.76 OR7E47Polfactory receptor, family 7, subfamily E, member 47 pseudogene (216698_x_at), score: -0.7 PARVBparvin, beta (37965_at), score: 0.79 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.72 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.62 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.71 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.69 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.69 PDE7Bphosphodiesterase 7B (220343_at), score: -0.74 PHF7PHD finger protein 7 (215622_x_at), score: -0.81 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.86 PKN1protein kinase N1 (202161_at), score: 0.76 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.74 PLK3polo-like kinase 3 (Drosophila) (204958_at), score: 0.61 PNPLA6patatin-like phospholipase domain containing 6 (203718_at), score: 0.66 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.69 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.62 PORP450 (cytochrome) oxidoreductase (208928_at), score: 0.69 PRKCDprotein kinase C, delta (202545_at), score: 0.79 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.74 PXNpaxillin (211823_s_at), score: 0.8 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.66 RAB22ARAB22A, member RAS oncogene family (218360_at), score: 0.6 RANBP3RAN binding protein 3 (210120_s_at), score: 0.63 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.63 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: 0.66 RLN1relaxin 1 (211753_s_at), score: -0.71 RNF130ring finger protein 130 (217865_at), score: 0.63 RNF24ring finger protein 24 (204669_s_at), score: 0.61 RNF40ring finger protein 40 (206845_s_at), score: 0.63 RP3-377H14.5hypothetical LOC285830 (222279_at), score: -0.73 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: -0.73 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.88 RPL21P68ribosomal protein L21 pseudogene 68 (217340_at), score: -0.7 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.85 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.72 SBF1SET binding factor 1 (39835_at), score: 0.6 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.7 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.94 SDAD1SDA1 domain containing 1 (218607_s_at), score: 0.7 SDC3syndecan 3 (202898_at), score: 0.65 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: -0.7 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.72 SHBSrc homology 2 domain containing adaptor protein B (204657_s_at), score: 0.67 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.71 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: -0.74 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: 0.71 SLC2A8solute carrier family 2 (facilitated glucose transporter), member 8 (218985_at), score: 0.62 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.79 SLC9A1solute carrier family 9 (sodium/hydrogen exchanger), member 1 (209453_at), score: 0.65 SLC9A3R2solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 (209830_s_at), score: 0.68 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.77 SMYD5SMYD family member 5 (209516_at), score: 0.62 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: 0.61 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.78 SPATA20spermatogenesis associated 20 (218164_at), score: 0.63 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.67 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: 0.62 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: 0.71 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.75 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.8 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.62 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.63 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.71 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.62 TGM4transglutaminase 4 (prostate) (217566_s_at), score: -0.7 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.71 TLE6transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) (222219_s_at), score: -0.71 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.78 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.74 TSKUtsukushin (218245_at), score: 0.73 TXLNAtaxilin alpha (212300_at), score: 0.63 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.87 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.61 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: -0.76 VEGFBvascular endothelial growth factor B (203683_s_at), score: 1 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.83 WBP2WW domain binding protein 2 (209117_at), score: 0.64 WDR42AWD repeat domain 42A (202249_s_at), score: 0.75 WDR6WD repeat domain 6 (217734_s_at), score: 0.76 XAB2XPA binding protein 2 (218110_at), score: 0.61 ZFHX3zinc finger homeobox 3 (208033_s_at), score: 0.67 ZNF282zinc finger protein 282 (212892_at), score: 0.61
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |