
Under-expression is coded with green,
over-expression with red color.

reproduction
The production by an organism of new individuals that contain some portion of their genetic material inherited from that organism.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
cellular alcohol metabolic process
The chemical reactions and pathways involving alcohols, any of a class of compounds containing one or more hydroxyl groups attached to a saturated carbon atom, as carried out by individual cells.
lipid metabolic process
The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism.
lipid transport
The directed movement of lipids into, out of, within or between cells. Lipids are compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
female pregnancy
The set of physiological processes that allow an embryo or foetus to develop within the body of a female animal. It covers the time from fertilization of a female ovum by a male spermatozoon until birth.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
steroid metabolic process
The chemical reactions and pathways involving steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus.
cholesterol metabolic process
The chemical reactions and pathways involving cholesterol, cholest-5-en-3 beta-ol, the principal sterol of vertebrates and the precursor of many steroids, including bile acids and steroid hormones. It is a component of the plasma membrane lipid bilayer and of plasma lipoproteins and can be found in all animal tissues.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
phospholipid transport
The directed movement of phospholipids into, out of, within or between cells. Phospholipids are any lipids containing phosphoric acid as a mono- or diester.
sterol metabolic process
The chemical reactions and pathways involving sterols, steroids with one or more hydroxyl groups and a hydrocarbon side-chain in the molecule.
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
phospholipid efflux
The directed movement of a phospholipid out of a cell or organelle.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular lipid metabolic process
The chemical reactions and pathways involving lipids, as carried out by individual cells.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
multi-organism process
Any process by which an organism has an effect on another organism of the same or different species.
all
This term is the most general term possible
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
female pregnancy
The set of physiological processes that allow an embryo or foetus to develop within the body of a female animal. It covers the time from fertilization of a female ovum by a male spermatozoon until birth.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
steroid metabolic process
The chemical reactions and pathways involving steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus.
cellular lipid metabolic process
The chemical reactions and pathways involving lipids, as carried out by individual cells.
sterol metabolic process
The chemical reactions and pathways involving sterols, steroids with one or more hydroxyl groups and a hydrocarbon side-chain in the molecule.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
receptor binding
Interacting selectively with one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function.
cytokine activity
Functions to control the survival, growth, differentiation and effector function of tissues and cells.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
all
This term is the most general term possible
ABCA1ATP-binding cassette, sub-family A (ABC1), member 1 (203504_s_at), score: 0.57 ABCA8ATP-binding cassette, sub-family A (ABC1), member 8 (204719_at), score: 0.6 ABCG1ATP-binding cassette, sub-family G (WHITE), member 1 (204567_s_at), score: 0.7 ACTBactin, beta (AFFX-HSAC07/X00351_5_at), score: -0.46 ACTR2ARP2 actin-related protein 2 homolog (yeast) (200727_s_at), score: -0.48 ADFPadipose differentiation-related protein (209122_at), score: 0.55 ADIPOR2adiponectin receptor 2 (201346_at), score: -0.47 AP3B1adaptor-related protein complex 3, beta 1 subunit (203142_s_at), score: -0.44 APOC1apolipoprotein C-I (213553_x_at), score: 0.57 ARAP3ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 (218950_at), score: 0.69 AREGamphiregulin (205239_at), score: 0.65 ARHGAP24Rho GTPase activating protein 24 (221030_s_at), score: 0.6 ARHGAP6Rho GTPase activating protein 6 (206167_s_at), score: 0.64 ARPC2actin related protein 2/3 complex, subunit 2, 34kDa (208679_s_at), score: -0.5 ARPC5Lactin related protein 2/3 complex, subunit 5-like (220966_x_at), score: -0.53 ASAP1ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 (221039_s_at), score: -0.48 ATF7IP2activating transcription factor 7 interacting protein 2 (219870_at), score: 0.52 AZI25-azacytidine induced 2 (218043_s_at), score: 0.75 BACH2BTB and CNC homology 1, basic leucine zipper transcription factor 2 (221234_s_at), score: 0.54 BCL11AB-cell CLL/lymphoma 11A (zinc finger protein) (219497_s_at), score: 0.87 BDKRB2bradykinin receptor B2 (205870_at), score: 0.71 BGNbiglycan (213905_x_at), score: 0.64 C10orf26chromosome 10 open reading frame 26 (202808_at), score: 0.59 C16orf7chromosome 16 open reading frame 7 (205781_at), score: 0.54 C1QTNF1C1q and tumor necrosis factor related protein 1 (220975_s_at), score: 0.64 C3complement component 3 (217767_at), score: 0.9 C5orf4chromosome 5 open reading frame 4 (48031_r_at), score: 0.74 C6orf66chromosome 6 open reading frame 66 (219006_at), score: -0.46 C9orf7chromosome 9 open reading frame 7 (61874_at), score: 0.55 CAMK2Bcalcium/calmodulin-dependent protein kinase II beta (34846_at), score: 0.54 CAPN3calpain 3, (p94) (210944_s_at), score: 0.54 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: 0.74 CCL7chemokine (C-C motif) ligand 7 (208075_s_at), score: 0.75 CCNG2cyclin G2 (202769_at), score: 0.6 CD44CD44 molecule (Indian blood group) (210916_s_at), score: -0.5 CD59CD59 molecule, complement regulatory protein (200984_s_at), score: -0.43 CEBPBCCAAT/enhancer binding protein (C/EBP), beta (212501_at), score: 0.52 CH25Hcholesterol 25-hydroxylase (206932_at), score: 1 CHI3L1chitinase 3-like 1 (cartilage glycoprotein-39) (209395_at), score: 0.81 COL16A1collagen, type XVI, alpha 1 (204345_at), score: 0.53 CPEcarboxypeptidase E (201117_s_at), score: 0.64 CPEB3cytoplasmic polyadenylation element binding protein 3 (205773_at), score: 0.58 CREB5cAMP responsive element binding protein 5 (205931_s_at), score: 0.57 CSF1colony stimulating factor 1 (macrophage) (209716_at), score: -0.45 CSNK1A1casein kinase 1, alpha 1 (206562_s_at), score: -0.48 CTAGE5CTAGE family, member 5 (215930_s_at), score: 0.61 CTNScystinosis, nephropathic (36566_at), score: 0.53 CXCL5chemokine (C-X-C motif) ligand 5 (214974_x_at), score: 0.79 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: 0.58 DDX21DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 (208152_s_at), score: -0.48 DHCR2424-dehydrocholesterol reductase (200862_at), score: -0.51 DHX15DEAH (Asp-Glu-Ala-His) box polypeptide 15 (201386_s_at), score: -0.49 DKC1dyskeratosis congenita 1, dyskerin (201478_s_at), score: -0.5 DKFZP586H2123regeneration associated muscle protease (213661_at), score: 0.58 DLG4discs, large homolog 4 (Drosophila) (204592_at), score: 0.52 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: 0.6 DNTTIP2deoxynucleotidyltransferase, terminal, interacting protein 2 (202776_at), score: -0.5 DOCK4dedicator of cytokinesis 4 (205003_at), score: 0.52 DUSP14dual specificity phosphatase 14 (203367_at), score: -0.43 EDNRAendothelin receptor type A (204464_s_at), score: 0.61 EIF4G1eukaryotic translation initiation factor 4 gamma, 1 (208624_s_at), score: -0.43 ELOVL4elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 (219532_at), score: 0.53 ETF1eukaryotic translation termination factor 1 (201574_at), score: -0.44 ETV1ets variant 1 (221911_at), score: 0.61 EXOSC9exosome component 9 (205061_s_at), score: -0.43 FCGRTFc fragment of IgG, receptor, transporter, alpha (218831_s_at), score: 0.59 FHITfragile histidine triad gene (206492_at), score: 0.62 FLJ10404hypothetical protein FLJ10404 (218920_at), score: 0.57 FMODfibromodulin (202709_at), score: 0.87 FTH1ferritin, heavy polypeptide 1 (214211_at), score: 0.6 FTHP1ferritin, heavy polypeptide pseudogene 1 (211628_x_at), score: 0.55 FUSIP1FUS interacting protein (serine/arginine-rich) 1 (210178_x_at), score: -0.45 GAP43growth associated protein 43 (204471_at), score: 0.56 GAS7growth arrest-specific 7 (202191_s_at), score: 0.64 GDF15growth differentiation factor 15 (221577_x_at), score: 0.75 GOLGA8Agolgi autoantigen, golgin subfamily a, 8A (208798_x_at), score: 0.57 GOLSYNGolgi-localized protein (218692_at), score: 0.62 GPM6Bglycoprotein M6B (209167_at), score: 0.83 GPR153G protein-coupled receptor 153 (221902_at), score: 0.53 H6PDhexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) (221892_at), score: 0.6 HBP1HMG-box transcription factor 1 (209102_s_at), score: 0.56 HEPHhephaestin (203903_s_at), score: 0.66 HIRAHIR histone cell cycle regulation defective homolog A (S. cerevisiae) (217427_s_at), score: -0.48 HIST3H2Ahistone cluster 3, H2a (221582_at), score: 0.59 HMGCR3-hydroxy-3-methylglutaryl-Coenzyme A reductase (202539_s_at), score: -0.47 HNRNPRheterogeneous nuclear ribonucleoprotein R (208765_s_at), score: -0.46 HSD11B1hydroxysteroid (11-beta) dehydrogenase 1 (205404_at), score: 0.68 HSPA14heat shock 70kDa protein 14 (219212_at), score: -0.45 HSPBAP1HSPB (heat shock 27kDa) associated protein 1 (219284_at), score: 0.57 IDI1isopentenyl-diphosphate delta isomerase 1 (204615_x_at), score: -0.63 IGFBP2insulin-like growth factor binding protein 2, 36kDa (202718_at), score: 0.63 IL1Binterleukin 1, beta (39402_at), score: 0.66 IL1RNinterleukin 1 receptor antagonist (212657_s_at), score: 0.92 IL24interleukin 24 (206569_at), score: 0.88 ING1inhibitor of growth family, member 1 (208415_x_at), score: 0.69 INSIG2insulin induced gene 2 (209566_at), score: 0.53 INSRinsulin receptor (213792_s_at), score: 0.52 ITGA10integrin, alpha 10 (206766_at), score: 0.54 JAM2junctional adhesion molecule 2 (219213_at), score: 0.64 KANK1KN motif and ankyrin repeat domains 1 (213005_s_at), score: -0.58 KCNMB4potassium large conductance calcium-activated channel, subfamily M, beta member 4 (219287_at), score: 0.56 KIAA0240KIAA0240 (38892_at), score: 0.52 KIAA0644KIAA0644 gene product (205150_s_at), score: 0.7 KPNA3karyopherin alpha 3 (importin alpha 4) (221503_s_at), score: -0.51 LOC391132similar to hCG2041276 (216177_at), score: 0.56 LRRC32leucine rich repeat containing 32 (203835_at), score: 0.72 LSG1large subunit GTPase 1 homolog (S. cerevisiae) (221535_at), score: -0.45 LYNv-yes-1 Yamaguchi sarcoma viral related oncogene homolog (202625_at), score: -0.48 MACROD1MACRO domain containing 1 (219188_s_at), score: 0.57 MAGOHmago-nashi homolog, proliferation-associated (Drosophila) (210092_at), score: -0.43 MAP2K3mitogen-activated protein kinase kinase 3 (207667_s_at), score: -0.49 MAP7microtubule-associated protein 7 (202890_at), score: 0.56 MAPRE1microtubule-associated protein, RP/EB family, member 1 (200712_s_at), score: -0.45 MCFD2multiple coagulation factor deficiency 2 (212246_at), score: -0.45 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: 0.52 MMEmembrane metallo-endopeptidase (203434_s_at), score: 0.52 MRPL35mitochondrial ribosomal protein L35 (218890_x_at), score: -0.46 MUC3Amucin 3A, cell surface associated (217117_x_at), score: 0.53 MXI1MAX interactor 1 (202364_at), score: 0.56 MYBL1v-myb myeloblastosis viral oncogene homolog (avian)-like 1 (213906_at), score: -0.44 NAV2neuron navigator 2 (218330_s_at), score: -0.46 NIP7nuclear import 7 homolog (S. cerevisiae) (219031_s_at), score: -0.51 NOTCH2Notch homolog 2 (Drosophila) (210756_s_at), score: -0.51 NOVA1neuro-oncological ventral antigen 1 (205794_s_at), score: 0.57 NOX4NADPH oxidase 4 (219773_at), score: 0.56 NPAS2neuronal PAS domain protein 2 (39549_at), score: 0.52 NR3C1nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) (201866_s_at), score: -0.59 NR4A2nuclear receptor subfamily 4, group A, member 2 (216248_s_at), score: 0.85 ODZ4odz, odd Oz/ten-m homolog 4 (Drosophila) (213273_at), score: 0.61 OSGIN2oxidative stress induced growth inhibitor family member 2 (204024_at), score: 0.54 PAPPApregnancy-associated plasma protein A, pappalysin 1 (201981_at), score: 0.58 PCSK5proprotein convertase subtilisin/kexin type 5 (205559_s_at), score: 0.56 PCYOX1prenylcysteine oxidase 1 (203803_at), score: -0.46 PDPNpodoplanin (221898_at), score: 0.83 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: 0.66 PERLD1per1-like domain containing 1 (55616_at), score: 0.55 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: 0.55 PLA2R1phospholipase A2 receptor 1, 180kDa (207415_at), score: 0.65 PLAG1pleiomorphic adenoma gene 1 (205372_at), score: 0.57 PLCL2phospholipase C-like 2 (213309_at), score: 0.52 PLEKHB2pleckstrin homology domain containing, family B (evectins) member 2 (201411_s_at), score: -0.45 PLGLB2plasminogen-like B2 (205871_at), score: 0.55 PLTPphospholipid transfer protein (202075_s_at), score: 0.6 POLR1Cpolymerase (RNA) I polypeptide C, 30kDa (207515_s_at), score: -0.43 PON2paraoxonase 2 (210830_s_at), score: -0.46 PSD3pleckstrin and Sec7 domain containing 3 (203355_s_at), score: 0.58 PSMD6proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 (202753_at), score: -0.44 PSME4proteasome (prosome, macropain) activator subunit 4 (212220_at), score: -0.45 PTGESprostaglandin E synthase (210367_s_at), score: 0.75 PTHLHparathyroid hormone-like hormone (211756_at), score: 0.62 PTPRDprotein tyrosine phosphatase, receptor type, D (214043_at), score: 0.96 PTPRNprotein tyrosine phosphatase, receptor type, N (204945_at), score: 0.62 PTPRUprotein tyrosine phosphatase, receptor type, U (211320_s_at), score: 0.78 QPCTglutaminyl-peptide cyclotransferase (205174_s_at), score: 0.55 RAB5CRAB5C, member RAS oncogene family (201156_s_at), score: -0.49 RBM14RNA binding motif protein 14 (204178_s_at), score: -0.44 RGS17regulator of G-protein signaling 17 (220334_at), score: 0.56 RGS5regulator of G-protein signaling 5 (218353_at), score: 0.67 RNF115ring finger protein 115 (212742_at), score: -0.47 RP11-345P4.4similar to solute carrier family 35, member E2 (217122_s_at), score: 0.52 RP4-691N24.1ninein-like (207705_s_at), score: 0.55 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: 0.57 RPS27Aribosomal protein S27a (217144_at), score: 0.57 RRAS2related RAS viral (r-ras) oncogene homolog 2 (208456_s_at), score: -0.58 S100A10S100 calcium binding protein A10 (200872_at), score: -0.43 SALL2sal-like 2 (Drosophila) (213283_s_at), score: 0.53 SEMA4Gsema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G (219194_at), score: 0.53 SERPINF1serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 (202283_at), score: 0.56 SERPING1serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 (200986_at), score: 0.62 SESN1sestrin 1 (218346_s_at), score: 0.52 SFRP4secreted frizzled-related protein 4 (204051_s_at), score: 0.59 SFRS7splicing factor, arginine/serine-rich 7, 35kDa (214141_x_at), score: -0.45 SLC22A17solute carrier family 22, member 17 (218675_at), score: 0.75 SLC39A14solute carrier family 39 (zinc transporter), member 14 (212110_at), score: 0.56 SLC7A7solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 (204588_s_at), score: 0.78 SMOXspermine oxidase (210357_s_at), score: 0.57 SNED1sushi, nidogen and EGF-like domains 1 (213488_at), score: 0.6 SNRPA1small nuclear ribonucleoprotein polypeptide A' (216977_x_at), score: -0.53 SPRY1sprouty homolog 1, antagonist of FGF signaling (Drosophila) (212558_at), score: 0.63 SPSB3splA/ryanodine receptor domain and SOCS box containing 3 (46256_at), score: 0.57 SSR4signal sequence receptor, delta (translocon-associated protein delta) (201004_at), score: 0.58 STIP1stress-induced-phosphoprotein 1 (212009_s_at), score: -0.46 STRA6stimulated by retinoic acid gene 6 homolog (mouse) (221701_s_at), score: 0.75 TDO2tryptophan 2,3-dioxygenase (205943_at), score: 0.63 TIMM23translocase of inner mitochondrial membrane 23 homolog (yeast) (218119_at), score: -0.52 TJP2tight junction protein 2 (zona occludens 2) (202085_at), score: -0.56 TMEM161Atransmembrane protein 161A (218745_x_at), score: 0.55 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.57 TPCN1two pore segment channel 1 (217914_at), score: 0.6 TPD52tumor protein D52 (201690_s_at), score: -0.45 TRNAG6transfer RNA glycine 6 (anticodon GCC) (217542_at), score: 0.58 TRPV1transient receptor potential cation channel, subfamily V, member 1 (219632_s_at), score: 0.56 UCK2uridine-cytidine kinase 2 (209825_s_at), score: -0.64 UGT2B28UDP glucuronosyltransferase 2 family, polypeptide B28 (211682_x_at), score: 0.52 WBP11WW domain binding protein 11 (217821_s_at), score: -0.44 WDTC1WD and tetratricopeptide repeats 1 (40829_at), score: 0.76 WIPI1WD repeat domain, phosphoinositide interacting 1 (203827_at), score: 0.53 WNT5Awingless-type MMTV integration site family, member 5A (213425_at), score: 0.53 YIPF5Yip1 domain family, member 5 (221423_s_at), score: -0.45 YWHAZtyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (200641_s_at), score: -0.5 ZFYVE16zinc finger, FYVE domain containing 16 (203651_at), score: 0.55 ZNF467zinc finger protein 467 (214746_s_at), score: 0.53
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
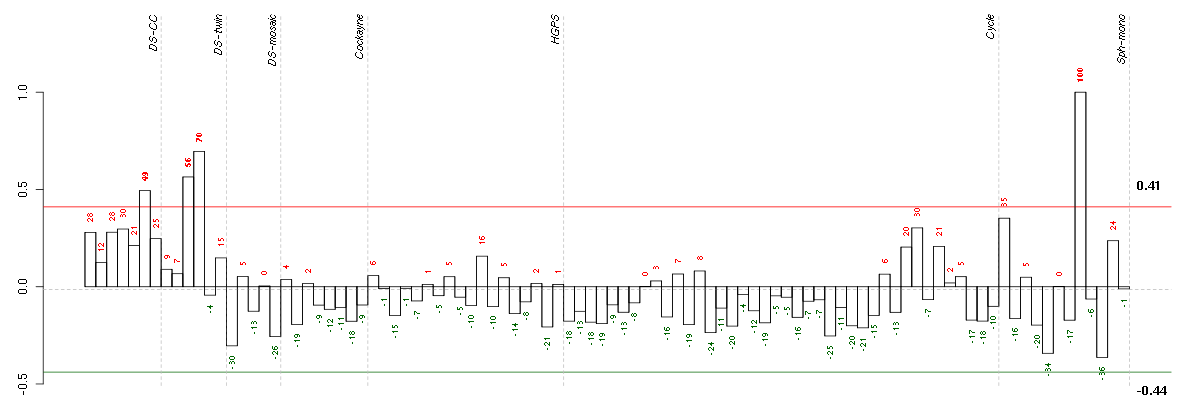
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |