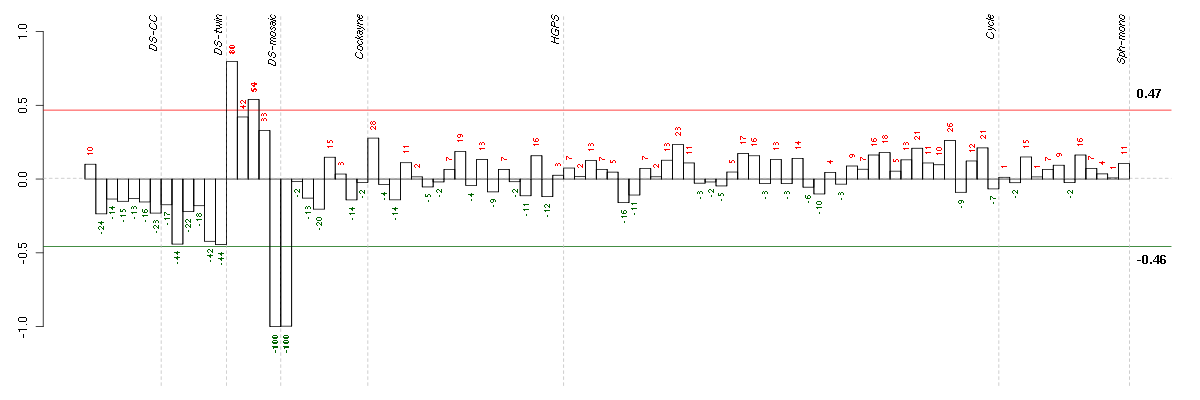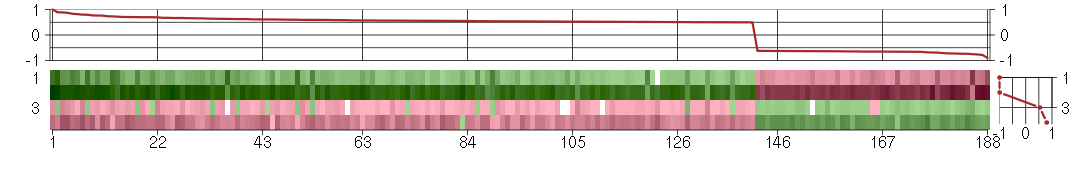

Under-expression is coded with green,
over-expression with red color.

biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

ADAM23ADAM metallopeptidase domain 23 (206046_at), score: 0.71 ADAMTS12ADAM metallopeptidase with thrombospondin type 1 motif, 12 (221421_s_at), score: -0.69 ADAP1ArfGAP with dual PH domains 1 (90265_at), score: 0.56 ADMadrenomedullin (202912_at), score: -0.68 AFPalpha-fetoprotein (204694_at), score: -0.65 AGRNagrin (212285_s_at), score: 0.67 AK3L1adenylate kinase 3-like 1 (204348_s_at), score: -0.73 ALDOBaldolase B, fructose-bisphosphate (214423_x_at), score: -0.63 ALG3asparagine-linked glycosylation 3, alpha-1,3- mannosyltransferase homolog (S. cerevisiae) (207396_s_at), score: 0.55 APOL1apolipoprotein L, 1 (209546_s_at), score: 0.63 ARID5AAT rich interactive domain 5A (MRF1-like) (213138_at), score: 0.61 ARSAarylsulfatase A (204443_at), score: 0.56 ASCC2activating signal cointegrator 1 complex subunit 2 (215684_s_at), score: 0.52 ATP5OATP synthase, H+ transporting, mitochondrial F1 complex, O subunit (216954_x_at), score: -0.63 B4GALNT1beta-1,4-N-acetyl-galactosaminyl transferase 1 (206435_at), score: -0.64 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.56 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: 0.64 BCRbreakpoint cluster region (202315_s_at), score: 0.59 BTN2A2butyrophilin, subfamily 2, member A2 (205298_s_at), score: -0.63 C19orf28chromosome 19 open reading frame 28 (220178_at), score: 0.56 C6orf105chromosome 6 open reading frame 105 (215100_at), score: -0.62 CA11carbonic anhydrase XI (209726_at), score: 0.83 CASP4caspase 4, apoptosis-related cysteine peptidase (209310_s_at), score: 0.52 CC2D1Acoiled-coil and C2 domain containing 1A (58994_at), score: 0.6 CFIcomplement factor I (203854_at), score: 0.54 CLDN1claudin 1 (218182_s_at), score: 0.69 CNN1calponin 1, basic, smooth muscle (203951_at), score: 0.52 COG2component of oligomeric golgi complex 2 (203073_at), score: 0.52 COL15A1collagen, type XV, alpha 1 (203477_at), score: 0.6 COL4A5collagen, type IV, alpha 5 (213110_s_at), score: 0.53 COQ4coenzyme Q4 homolog (S. cerevisiae) (218328_at), score: 0.5 CORO1Bcoronin, actin binding protein, 1B (64486_at), score: 0.52 CRIP2cysteine-rich protein 2 (208978_at), score: 0.58 CUX1cut-like homeobox 1 (214743_at), score: 0.5 CYP2B7P1cytochrome P450, family 2, subfamily B, polypeptide 7 pseudogene 1 (210272_at), score: -0.63 DNAJB5DnaJ (Hsp40) homolog, subfamily B, member 5 (212817_at), score: 0.56 DNAJC4DnaJ (Hsp40) homolog, subfamily C, member 4 (206782_s_at), score: 0.52 DNASE2deoxyribonuclease II, lysosomal (214992_s_at), score: 0.54 DOK5docking protein 5 (214844_s_at), score: 0.5 DPYSL4dihydropyrimidinase-like 4 (205493_s_at), score: 0.53 DYNLT3dynein, light chain, Tctex-type 3 (203303_at), score: -0.74 ECE1endothelin converting enzyme 1 (201750_s_at), score: 0.56 ELOVL2elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 (213712_at), score: 0.72 ELSPBP1epididymal sperm binding protein 1 (220366_at), score: -0.66 ENPP4ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative function) (204160_s_at), score: -0.72 ERAP1endoplasmic reticulum aminopeptidase 1 (214012_at), score: 0.8 ERN1endoplasmic reticulum to nucleus signaling 1 (207061_at), score: -0.62 FAM102Afamily with sequence similarity 102, member A (212400_at), score: 0.61 FHL3four and a half LIM domains 3 (218818_at), score: 0.56 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.57 FOSv-fos FBJ murine osteosarcoma viral oncogene homolog (209189_at), score: 0.89 FOXF1forkhead box F1 (205935_at), score: 0.62 FRAP1FK506 binding protein 12-rapamycin associated protein 1 (202288_at), score: 0.52 GALNT3UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) (203397_s_at), score: 0.61 GDF15growth differentiation factor 15 (221577_x_at), score: 0.54 GNG4guanine nucleotide binding protein (G protein), gamma 4 (205184_at), score: -0.72 GP1BBglycoprotein Ib (platelet), beta polypeptide (206655_s_at), score: 0.6 GPC4glypican 4 (204983_s_at), score: 0.59 GPR37G protein-coupled receptor 37 (endothelin receptor type B-like) (209631_s_at), score: 0.5 GSPT2G1 to S phase transition 2 (205541_s_at), score: 0.54 GTPBP1GTP binding protein 1 (219357_at), score: 0.53 HAPLN1hyaluronan and proteoglycan link protein 1 (205523_at), score: -0.65 HES1hairy and enhancer of split 1, (Drosophila) (203394_s_at), score: 0.54 HHEXhematopoietically expressed homeobox (215933_s_at), score: 0.82 HOXB9homeobox B9 (216417_x_at), score: -0.75 HOXC6homeobox C6 (206858_s_at), score: -0.74 HOXD1homeobox D1 (205975_s_at), score: -0.66 HSF1heat shock transcription factor 1 (202344_at), score: 0.68 ICAM1intercellular adhesion molecule 1 (202638_s_at), score: 0.56 IL1RAPinterleukin 1 receptor accessory protein (205227_at), score: 0.52 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: 0.8 ISYNA1inositol-3-phosphate synthase 1 (222240_s_at), score: 0.66 JUPjunction plakoglobin (201015_s_at), score: 0.7 KIAA0556KIAA0556 (213185_at), score: 0.52 KLHL26kelch-like 26 (Drosophila) (219354_at), score: 0.51 KNG1kininogen 1 (217512_at), score: -0.64 KRASv-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (214352_s_at), score: -0.66 KTELC1KTEL (Lys-Tyr-Glu-Leu) containing 1 (218587_s_at), score: 0.6 LAMA2laminin, alpha 2 (213519_s_at), score: -0.63 LAMA5laminin, alpha 5 (210150_s_at), score: 0.5 LEPREL2leprecan-like 2 (204854_at), score: 0.51 LGALS3BPlectin, galactoside-binding, soluble, 3 binding protein (200923_at), score: 0.53 LILRA5leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 (215838_at), score: -0.65 LITAFlipopolysaccharide-induced TNF factor (200706_s_at), score: 0.56 LOC100170939glucuronidase, beta pseudogene (214850_at), score: -0.65 LRP5low density lipoprotein receptor-related protein 5 (209468_at), score: 0.67 MANBAmannosidase, beta A, lysosomal (203778_at), score: 0.54 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.51 MAP3K11mitogen-activated protein kinase kinase kinase 11 (203652_at), score: 0.59 MCCC1methylcrotonoyl-Coenzyme A carboxylase 1 (alpha) (218440_at), score: 0.51 ME3malic enzyme 3, NADP(+)-dependent, mitochondrial (204663_at), score: 0.54 MMRN2multimerin 2 (219091_s_at), score: -0.71 MRPS2mitochondrial ribosomal protein S2 (218001_at), score: 0.51 MSR1macrophage scavenger receptor 1 (208423_s_at), score: -0.78 MTSS1metastasis suppressor 1 (203037_s_at), score: 0.63 MUC3Bmucin 3B, cell surface associated (214898_x_at), score: 0.54 MYBBP1AMYB binding protein (P160) 1a (219098_at), score: 0.53 MYO1Amyosin IA (211916_s_at), score: -0.62 NCAM2neural cell adhesion molecule 2 (205669_at), score: 0.5 NCDNneurochondrin (209556_at), score: 0.5 NCRNA00084non-protein coding RNA 84 (214657_s_at), score: -0.64 NESnestin (218678_at), score: 0.55 NINJ1ninjurin 1 (203045_at), score: 0.73 NXPH4neurexophilin 4 (221967_at), score: 0.59 OS9osteosarcoma amplified 9, endoplasmic reticulum associated protein (200714_x_at), score: 0.51 P2RX4purinergic receptor P2X, ligand-gated ion channel, 4 (204088_at), score: 0.56 PAPPA2pappalysin 2 (213332_at), score: 0.73 PARP11poly (ADP-ribose) polymerase family, member 11 (220315_at), score: -0.66 PCBD1pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (203557_s_at), score: 0.61 PCDH7protocadherin 7 (205534_at), score: 0.53 PDCD11programmed cell death 11 (212422_at), score: 0.5 PDE1Cphosphodiesterase 1C, calmodulin-dependent 70kDa (207303_at), score: -0.9 PDE4Aphosphodiesterase 4A, cAMP-specific (phosphodiesterase E2 dunce homolog, Drosophila) (204735_at), score: 0.56 PDGFAplatelet-derived growth factor alpha polypeptide (205463_s_at), score: 0.51 PDK2pyruvate dehydrogenase kinase, isozyme 2 (202590_s_at), score: 0.55 PDLIM3PDZ and LIM domain 3 (209621_s_at), score: 0.77 PHKA2phosphorylase kinase, alpha 2 (liver) (209439_s_at), score: 0.57 PHLDA3pleckstrin homology-like domain, family A, member 3 (218634_at), score: 0.5 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.64 PKP2plakophilin 2 (207717_s_at), score: 0.89 PLAGL2pleiomorphic adenoma gene-like 2 (202925_s_at), score: 0.65 PLTPphospholipid transfer protein (202075_s_at), score: 0.57 PLXNB1plexin B1 (215807_s_at), score: 0.63 PNMAL1PNMA-like 1 (218824_at), score: 0.59 PORP450 (cytochrome) oxidoreductase (208928_at), score: 0.5 PPIGpeptidylprolyl isomerase G (cyclophilin G) (208993_s_at), score: -0.63 PRKAB2protein kinase, AMP-activated, beta 2 non-catalytic subunit (214474_at), score: -0.63 PSG1pregnancy specific beta-1-glycoprotein 1 (208257_x_at), score: -0.69 PSG3pregnancy specific beta-1-glycoprotein 3 (215821_x_at), score: -0.76 PSG7pregnancy specific beta-1-glycoprotein 7 (205602_x_at), score: -0.65 PTGDSprostaglandin D2 synthase 21kDa (brain) (212187_x_at), score: 0.7 RAB33ARAB33A, member RAS oncogene family (206039_at), score: 0.54 RAB35RAB35, member RAS oncogene family (221819_at), score: 0.51 RAMP1receptor (G protein-coupled) activity modifying protein 1 (204916_at), score: 0.61 RANBP3RAN binding protein 3 (210120_s_at), score: 0.51 RARRES3retinoic acid receptor responder (tazarotene induced) 3 (204070_at), score: 0.63 RBM14RNA binding motif protein 14 (204178_s_at), score: 0.57 RGS2regulator of G-protein signaling 2, 24kDa (202388_at), score: 0.53 RHBDF2rhomboid 5 homolog 2 (Drosophila) (219202_at), score: 0.55 ROGDIrogdi homolog (Drosophila) (218394_at), score: 0.52 RP11-35N6.1plasticity related gene 3 (219732_at), score: -0.65 RRP1ribosomal RNA processing 1 homolog (S. cerevisiae) (218758_s_at), score: 0.55 SCARB1scavenger receptor class B, member 1 (201819_at), score: 0.87 SCG2secretogranin II (chromogranin C) (204035_at), score: 1 SEMA6Dsema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D (220574_at), score: -0.63 SENP5SUMO1/sentrin specific peptidase 5 (213184_at), score: 0.7 SGPL1sphingosine-1-phosphate lyase 1 (212322_at), score: 0.57 SHOX2short stature homeobox 2 (210135_s_at), score: -0.64 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.53 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: 0.59 SLC12A7solute carrier family 12 (potassium/chloride transporters), member 7 (218066_at), score: 0.63 SLC24A6solute carrier family 24 (sodium/potassium/calcium exchanger), member 6 (218749_s_at), score: 0.56 SLC25A22solute carrier family 25 (mitochondrial carrier: glutamate), member 22 (218725_at), score: 0.52 SLC2A8solute carrier family 2 (facilitated glucose transporter), member 8 (218985_at), score: 0.5 SLC9A1solute carrier family 9 (sodium/hydrogen exchanger), member 1 (209453_at), score: 0.67 SLCO1C1solute carrier organic anion transporter family, member 1C1 (220460_at), score: -0.63 SMARCD3SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 (204099_at), score: 0.58 SMG1SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) (210057_at), score: -0.64 SMOsmoothened homolog (Drosophila) (218629_at), score: 0.63 SPAG4sperm associated antigen 4 (219888_at), score: -0.65 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: 0.66 SSH1slingshot homolog 1 (Drosophila) (221753_at), score: -0.64 ST6GALNAC5ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 (220979_s_at), score: -0.64 STAP2signal transducing adaptor family member 2 (221610_s_at), score: -0.66 SUOXsulfite oxidase (204067_at), score: 0.5 SYT1synaptotagmin I (203999_at), score: 0.52 TGM2transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) (201042_at), score: 0.58 TM6SF1transmembrane 6 superfamily member 1 (219892_at), score: 0.76 TMCO6transmembrane and coiled-coil domains 6 (221096_s_at), score: -0.65 TMEM132Atransmembrane protein 132A (218834_s_at), score: 0.76 TMEM222transmembrane protein 222 (221512_at), score: 0.56 TNFSF4tumor necrosis factor (ligand) superfamily, member 4 (207426_s_at), score: 0.7 TNIKTRAF2 and NCK interacting kinase (213107_at), score: 0.65 TOB2transducer of ERBB2, 2 (221496_s_at), score: 0.54 TOM1target of myb1 (chicken) (202807_s_at), score: 0.51 TP53I11tumor protein p53 inducible protein 11 (214667_s_at), score: 0.52 TPCN1two pore segment channel 1 (217914_at), score: 0.57 TPD52tumor protein D52 (201690_s_at), score: 0.67 TRIB2tribbles homolog 2 (Drosophila) (202478_at), score: 0.59 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.51 VCAM1vascular cell adhesion molecule 1 (203868_s_at), score: 0.61 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: -0.63 WDR91WD repeat domain 91 (218971_s_at), score: -0.65 WT1Wilms tumor 1 (206067_s_at), score: 0.55 YRDCyrdC domain containing (E. coli) (218647_s_at), score: 0.5 ZNF282zinc finger protein 282 (212892_at), score: 0.5 ZNF35zinc finger protein 35 (206096_at), score: 0.71 ZNF711zinc finger protein 711 (207781_s_at), score: 0.7
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 47C.CEL | 5 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 5 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |