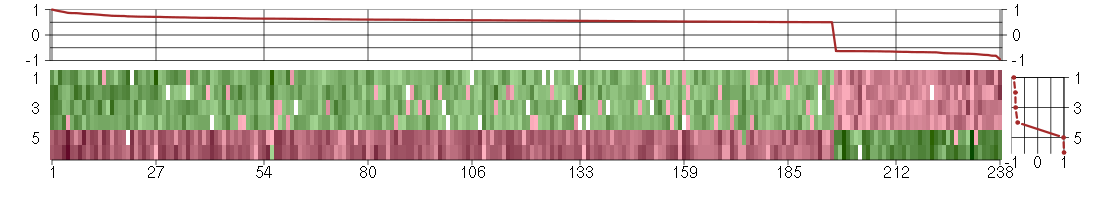

Under-expression is coded with green,
over-expression with red color.



ADARadenosine deaminase, RNA-specific (201786_s_at), score: 0.57 AGAP1ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 (204066_s_at), score: 0.64 AK5adenylate kinase 5 (219308_s_at), score: -0.64 AKIRIN1akirin 1 (217893_s_at), score: 0.58 ARL17P1ADP-ribosylation factor-like 17 pseudogene 1 (210718_s_at), score: 0.58 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: 0.71 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (221484_at), score: 0.66 BAHCC1BAH domain and coiled-coil containing 1 (219218_at), score: 0.51 BAIAP2BAI1-associated protein 2 (205294_at), score: 0.56 BIN1bridging integrator 1 (210201_x_at), score: 0.72 BRAFv-raf murine sarcoma viral oncogene homolog B1 (206044_s_at), score: 0.57 C11orf71chromosome 11 open reading frame 71 (218789_s_at), score: 0.55 C12orf49chromosome 12 open reading frame 49 (218867_s_at), score: 0.6 C14orf106chromosome 14 open reading frame 106 (206500_s_at), score: -0.81 C17orf86chromosome 17 open reading frame 86 (221621_at), score: 0.67 C19orf22chromosome 19 open reading frame 22 (221764_at), score: 0.61 C1orf103chromosome 1 open reading frame 103 (220235_s_at), score: 0.5 CBFA2T2core-binding factor, runt domain, alpha subunit 2; translocated to, 2 (207625_s_at), score: 0.57 CCDC47coiled-coil domain containing 47 (217814_at), score: 0.5 CCT6Bchaperonin containing TCP1, subunit 6B (zeta 2) (206587_at), score: 0.51 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: 0.59 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: 0.53 CDS2CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 (212864_at), score: 0.54 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: 0.52 CHKAcholine kinase alpha (204233_s_at), score: 0.8 CHPT1choline phosphotransferase 1 (221675_s_at), score: 0.58 CICcapicua homolog (Drosophila) (212784_at), score: 0.61 CISD3CDGSH iron sulfur domain 3 (213551_x_at), score: 0.58 CLCN6chloride channel 6 (203950_s_at), score: 0.59 COX10COX10 homolog, cytochrome c oxidase assembly protein, heme A: farnesyltransferase (yeast) (203858_s_at), score: 0.56 CREBZFCREB/ATF bZIP transcription factor (202978_s_at), score: 0.56 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.86 CTPS2CTP synthase II (219080_s_at), score: -0.66 CYorf15Bchromosome Y open reading frame 15B (214131_at), score: 0.52 CYTH1cytohesin 1 (202880_s_at), score: 0.72 DCTPP1dCTP pyrophosphatase 1 (218069_at), score: -0.66 DDX6DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 (204909_at), score: 0.6 DENND4BDENN/MADD domain containing 4B (202860_at), score: 0.64 DHX32DEAH (Asp-Glu-Ala-His) box polypeptide 32 (218198_at), score: 0.51 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: 0.73 DOK4docking protein 4 (209691_s_at), score: 0.63 DUSP3dual specificity phosphatase 3 (201536_at), score: 0.58 DVL3dishevelled, dsh homolog 3 (Drosophila) (201908_at), score: 0.55 E2F8E2F transcription factor 8 (219990_at), score: -0.63 EPB41L1erythrocyte membrane protein band 4.1-like 1 (212339_at), score: 0.71 EPHA3EPH receptor A3 (211164_at), score: 0.64 EPHA4EPH receptor A4 (206114_at), score: -0.76 ETF1eukaryotic translation termination factor 1 (201574_at), score: 0.51 FAM102Afamily with sequence similarity 102, member A (212400_at), score: 0.59 FAM134Cfamily with sequence similarity 134, member C (212697_at), score: 0.69 FAM168Bfamily with sequence similarity 168, member B (212017_at), score: 0.64 FBXO41F-box protein 41 (44040_at), score: 0.55 FBXW11F-box and WD repeat domain containing 11 (209455_at), score: 0.56 FKBPLFK506 binding protein like (219187_at), score: -0.72 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.75 FNBP4formin binding protein 4 (212232_at), score: 0.65 FOXJ3forkhead box J3 (206015_s_at), score: 0.63 FOXK2forkhead box K2 (203064_s_at), score: 1 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: 0.51 GANgigaxonin (220124_at), score: 0.62 GGNBP2gametogenetin binding protein 2 (218079_s_at), score: 0.52 GIGYF2GRB10 interacting GYF protein 2 (212260_at), score: 0.53 GINS3GINS complex subunit 3 (Psf3 homolog) (45633_at), score: -0.78 GIT1G protein-coupled receptor kinase interacting ArfGAP 1 (218030_at), score: 0.69 GJA1gap junction protein, alpha 1, 43kDa (201667_at), score: -0.74 GOLGA3golgi autoantigen, golgin subfamily a, 3 (202106_at), score: 0.53 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: 0.64 GPATCH8G patch domain containing 8 (212485_at), score: 0.6 GPBP1L1GC-rich promoter binding protein 1-like 1 (217877_s_at), score: 0.59 GTF2Bgeneral transcription factor IIB (208066_s_at), score: 0.54 GTF2F2general transcription factor IIF, polypeptide 2, 30kDa (209595_at), score: -0.65 GTPBP1GTP binding protein 1 (219357_at), score: 0.66 HABP4hyaluronan binding protein 4 (209818_s_at), score: 0.65 HCG8HLA complex group 8 (215985_at), score: 0.51 HDAC9histone deacetylase 9 (205659_at), score: 0.6 HIST1H2BEhistone cluster 1, H2be (208527_x_at), score: -0.63 HIST1H2BHhistone cluster 1, H2bh (208546_x_at), score: -0.73 HIST1H3Hhistone cluster 1, H3h (206110_at), score: 0.57 HMGCR3-hydroxy-3-methylglutaryl-Coenzyme A reductase (202539_s_at), score: 0.62 HPS4Hermansky-Pudlak syndrome 4 (54037_at), score: 0.53 IGFBP5insulin-like growth factor binding protein 5 (203424_s_at), score: -0.63 IMAASLC7A5 pseudogene (208118_x_at), score: 0.64 INSIG1insulin induced gene 1 (201627_s_at), score: 0.52 INSIG2insulin induced gene 2 (209566_at), score: -0.63 INTS7integrator complex subunit 7 (222250_s_at), score: -0.68 IPPKinositol 1,3,4,5,6-pentakisphosphate 2-kinase (219092_s_at), score: 0.67 ISOC1isochorismatase domain containing 1 (218170_at), score: -0.63 ITGA1integrin, alpha 1 (214660_at), score: -0.64 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: -0.68 KCNN2potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 (220116_at), score: -0.64 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: 0.85 KCTD2potassium channel tetramerisation domain containing 2 (212564_at), score: 0.6 KIF1Bkinesin family member 1B (209234_at), score: 0.77 KLHDC10kelch domain containing 10 (209256_s_at), score: 0.7 KLHL18kelch-like 18 (Drosophila) (212882_at), score: 0.53 KPNA6karyopherin alpha 6 (importin alpha 7) (212101_at), score: 0.5 LAMB4laminin, beta 4 (215516_at), score: 0.52 LGALS1lectin, galactoside-binding, soluble, 1 (201105_at), score: -0.63 LHFPL2lipoma HMGIC fusion partner-like 2 (212658_at), score: 0.58 LOC100128223hypothetical protein LOC100128223 (221264_s_at), score: 0.51 LOC128192similar to peptidyl-Pro cis trans isomerase (217346_at), score: -0.81 LOC388796hypothetical LOC388796 (65588_at), score: 0.54 LOC440345hypothetical protein LOC440345 (214984_at), score: 0.56 LOC51152melanoma antigen (220771_at), score: 0.67 LOC645139hypothetical LOC645139 (209064_x_at), score: 0.72 LOC729034similar to puromycin sensitive aminopeptidase (214107_x_at), score: 0.57 LRRTM4leucine rich repeat transmembrane neuronal 4 (220345_at), score: 0.6 LSM14BLSM14B, SCD6 homolog B (S. cerevisiae) (219653_at), score: 0.59 LZTFL1leucine zipper transcription factor-like 1 (218437_s_at), score: 0.52 MAP1LC3Bmicrotubule-associated protein 1 light chain 3 beta (208786_s_at), score: 0.58 MAP2K3mitogen-activated protein kinase kinase 3 (207667_s_at), score: 0.6 MAP3K14mitogen-activated protein kinase kinase kinase 14 (205192_at), score: 0.61 MAPKAPK5mitogen-activated protein kinase-activated protein kinase 5 (212871_at), score: 0.63 MARK4MAP/microtubule affinity-regulating kinase 4 (55065_at), score: 0.51 MC4Rmelanocortin 4 receptor (221467_at), score: -0.63 MCTP1multiple C2 domains, transmembrane 1 (220122_at), score: 0.55 MEF2Amyocyte enhancer factor 2A (214684_at), score: 0.5 MGRN1mahogunin, ring finger 1 (212576_at), score: 0.55 MTMR3myotubularin related protein 3 (202197_at), score: 0.54 MTUS1mitochondrial tumor suppressor 1 (212096_s_at), score: 0.87 NBPF1neuroblastoma breakpoint family, member 1 (212854_x_at), score: 0.52 NBPF12neuroblastoma breakpoint family, member 12 (213612_x_at), score: 0.56 NCOA6nuclear receptor coactivator 6 (208979_at), score: 0.6 NCRNA00115non-protein coding RNA 115 (220399_at), score: 0.83 NEDD4Lneural precursor cell expressed, developmentally down-regulated 4-like (212445_s_at), score: 0.62 NENFneuron derived neurotrophic factor (218407_x_at), score: 0.57 NOTCH2NLNotch homolog 2 (Drosophila) N-terminal like (214722_at), score: 0.51 NPC2Niemann-Pick disease, type C2 (200701_at), score: 0.64 NTRK3neurotrophic tyrosine kinase, receptor, type 3 (217033_x_at), score: 0.54 OR7E156Polfactory receptor, family 7, subfamily E, member 156 pseudogene (222327_x_at), score: -0.71 OTUD3OTU domain containing 3 (213216_at), score: 0.6 PAIP1poly(A) binding protein interacting protein 1 (208051_s_at), score: 0.65 PARNpoly(A)-specific ribonuclease (deadenylation nuclease) (203905_at), score: 0.54 PCGF2polycomb group ring finger 2 (203793_x_at), score: 0.82 PDCD2programmed cell death 2 (213581_at), score: 0.56 PDE4Aphosphodiesterase 4A, cAMP-specific (phosphodiesterase E2 dunce homolog, Drosophila) (204735_at), score: 0.61 PEX19peroxisomal biogenesis factor 19 (201707_at), score: 0.54 PEX26peroxisomal biogenesis factor 26 (219180_s_at), score: 0.56 PEX5peroxisomal biogenesis factor 5 (203244_at), score: 0.54 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: 0.52 PHF20L1PHD finger protein 20-like 1 (222133_s_at), score: 0.59 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.96 PLAGL2pleiomorphic adenoma gene-like 2 (202925_s_at), score: 0.69 PMS2CLPMS2 C-terminal like pseudogene (221206_at), score: 0.73 POLR3Fpolymerase (RNA) III (DNA directed) polypeptide F, 39 kDa (205218_at), score: 0.58 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.66 POM121CPOM121 membrane glycoprotein C (213360_s_at), score: 0.6 PPARGC1Aperoxisome proliferator-activated receptor gamma, coactivator 1 alpha (219195_at), score: 0.57 PPP1R8protein phosphatase 1, regulatory (inhibitor) subunit 8 (207830_s_at), score: 0.51 PPP2R5Aprotein phosphatase 2, regulatory subunit B', alpha isoform (202187_s_at), score: 0.61 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: 0.68 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.63 RAB11FIP5RAB11 family interacting protein 5 (class I) (210879_s_at), score: 0.64 RAB22ARAB22A, member RAS oncogene family (218360_at), score: 0.93 RAD52RAD52 homolog (S. cerevisiae) (205647_at), score: 0.78 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: 0.58 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.79 RGNEFRho-guanine nucleotide exchange factor (219610_at), score: 0.5 RNF19Bring finger protein 19B (213038_at), score: 0.5 RNF220ring finger protein 220 (219988_s_at), score: 0.61 RNMTRNA (guanine-7-) methyltransferase (202683_s_at), score: 0.57 RP11-138L21.1similar to cell recognition molecule CASPR3 (220436_at), score: 0.54 RPL10P16ribosomal protein L10 pseudogene 16 (217680_x_at), score: -0.65 RPL5ribosomal protein L5 (213689_x_at), score: -0.67 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.5 RSAD1radical S-adenosyl methionine domain containing 1 (218307_at), score: 0.9 SAP30LSAP30-like (219129_s_at), score: 0.53 SAPS2SAPS domain family, member 2 (202792_s_at), score: 0.6 SAPS3SAPS domain family, member 3 (217928_s_at), score: 0.61 SDC4syndecan 4 (202071_at), score: 0.55 SENP5SUMO1/sentrin specific peptidase 5 (213184_at), score: 0.67 SGSHN-sulfoglucosamine sulfohydrolase (35626_at), score: 0.68 SIRT5sirtuin (silent mating type information regulation 2 homolog) 5 (S. cerevisiae) (219185_at), score: -0.66 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.86 SLC16A6solute carrier family 16, member 6 (monocarboxylic acid transporter 7) (207038_at), score: 0.54 SLC17A5solute carrier family 17 (anion/sugar transporter), member 5 (221041_s_at), score: 0.56 SLC19A1solute carrier family 19 (folate transporter), member 1 (211576_s_at), score: -0.67 SLC25A28solute carrier family 25, member 28 (221432_s_at), score: 0.67 SMCR7LSmith-Magenis syndrome chromosome region, candidate 7-like (221516_s_at), score: 0.7 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.82 SNIP1Smad nuclear interacting protein 1 (219409_at), score: 0.57 SNRKSNF related kinase (209481_at), score: 0.54 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: 0.54 SPAG8sperm associated antigen 8 (206816_s_at), score: -0.64 SPATA2spermatogenesis associated 2 (204433_s_at), score: 0.75 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.52 STAU1staufen, RNA binding protein, homolog 1 (Drosophila) (207320_x_at), score: 0.69 STAU2staufen, RNA binding protein, homolog 2 (Drosophila) (204226_at), score: -0.78 STK3serine/threonine kinase 3 (STE20 homolog, yeast) (204068_at), score: -0.73 STX3syntaxin 3 (209238_at), score: 0.64 SUPT5Hsuppressor of Ty 5 homolog (S. cerevisiae) (201480_s_at), score: 0.63 SYNMsynemin, intermediate filament protein (212730_at), score: -0.67 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.75 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.62 THUMPD2THUMP domain containing 2 (219248_at), score: 0.56 TLE6transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) (222219_s_at), score: -0.63 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: -0.66 TMEM149transmembrane protein 149 (219690_at), score: -0.75 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: 0.67 TNFRSF10Ctumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain (206222_at), score: 0.51 TOMM34translocase of outer mitochondrial membrane 34 (201870_at), score: 0.51 TOP3Atopoisomerase (DNA) III alpha (214300_s_at), score: -0.73 TRAPPC10trafficking protein particle complex 10 (209412_at), score: 0.64 TRIM52tripartite motif-containing 52 (221897_at), score: 0.55 TRIT1tRNA isopentenyltransferase 1 (218617_at), score: 0.59 TSKUtsukushin (218245_at), score: 0.71 TTC13tetratricopeptide repeat domain 13 (219481_at), score: -0.7 TXLNAtaxilin alpha (212300_at), score: 0.55 UBAP1ubiquitin associated protein 1 (221490_at), score: 0.59 UBE2Zubiquitin-conjugating enzyme E2Z (217750_s_at), score: 0.53 UBXN6UBX domain protein 6 (220757_s_at), score: 0.51 UIMC1ubiquitin interaction motif containing 1 (220746_s_at), score: 0.57 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.66 UPP1uridine phosphorylase 1 (203234_at), score: 0.64 USF2upstream transcription factor 2, c-fos interacting (214879_x_at), score: 0.51 VAMP2vesicle-associated membrane protein 2 (synaptobrevin 2) (201557_at), score: 0.7 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.61 WASF1WAS protein family, member 1 (204165_at), score: -0.68 WDR26WD repeat domain 26 (218107_at), score: 0.53 WDR42AWD repeat domain 42A (202249_s_at), score: 0.71 WHAMML1WAS protein homolog associated with actin, golgi membranes and microtubules-like 1 (213908_at), score: 0.53 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.72 ZAKsterile alpha motif and leucine zipper containing kinase AZK (218833_at), score: -0.64 ZBTB39zinc finger and BTB domain containing 39 (205256_at), score: 0.59 ZCCHC14zinc finger, CCHC domain containing 14 (212655_at), score: 0.58 ZDHHC14zinc finger, DHHC-type containing 14 (219247_s_at), score: -0.95 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.5 ZFP106zinc finger protein 106 homolog (mouse) (217781_s_at), score: 0.75 ZMAT3zinc finger, matrin type 3 (219628_at), score: 0.57 ZMIZ1zinc finger, MIZ-type containing 1 (212124_at), score: 0.53 ZNF212zinc finger protein 212 (203985_at), score: 0.58 ZNF264zinc finger protein 264 (205917_at), score: 0.58 ZNF432zinc finger protein 432 (219848_s_at), score: 0.61 ZNF544zinc finger protein 544 (218735_s_at), score: -0.67 ZNF652zinc finger protein 652 (205594_at), score: 0.52 ZNF706zinc finger protein 706 (218059_at), score: 0.53 ZSCAN12zinc finger and SCAN domain containing 12 (206507_at), score: 0.58
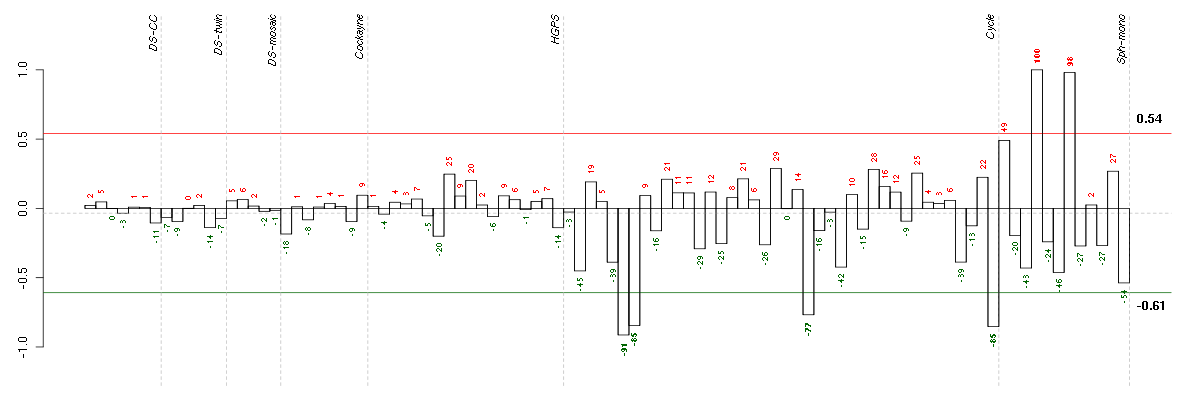
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |