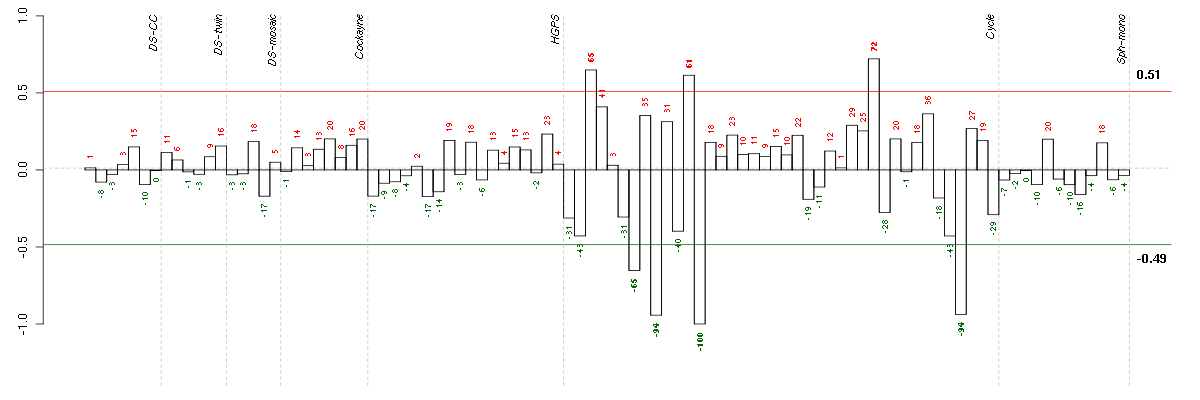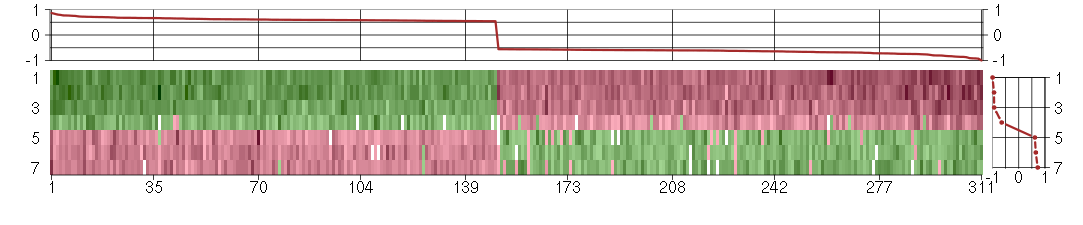

Under-expression is coded with green,
over-expression with red color.

protein import into nucleus
The directed movement of a protein from the cytoplasm to the nucleus.
protein localization
Any process by which a protein is transported to, or maintained in, a specific location.
protein targeting
The process of targeting specific proteins to particular membrane-bounded subcellular organelles. Usually requires an organelle specific protein sequence motif.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism.
intracellular protein transport
The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell.
nucleocytoplasmic transport
The directed movement of molecules between the nucleus and the cytoplasm.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
protein transport
The directed movement of proteins into, out of, within or between cells.
protein import
The directed movement of proteins into a cell or organelle.
regulation of intracellular transport
Any process that modulates the frequency, rate or extent of the directed movement of substances within cells.
negative regulation of intracellular transport
Any process that stops, prevents, or reduces the frequency, rate or extent of the directed movement of substances within cells.
regulation of localization
Any process that modulates the frequency, rate or extent of any process by which a cell, a substance, or a cellular entity is transported to, or maintained in, a specific location.
regulation of protein localization
Any process that modulates the frequency, rate or extent of any process by which a protein is transported to, or maintained in, a specific location.
macromolecule localization
Any process by which a macromolecule is transported to, or maintained in, a specific location.
regulation of intracellular protein transport
Any process that modulates the frequency, rate or extent of the directed movement of proteins within cells.
cellular protein localization
Any process by which a protein is transported to, and/or maintained in, a specific location within or at the surface of a cell.
regulation of protein import into nucleus
Any process that modulates the frequency, rate or extent of movement of proteins from the cytoplasm to the nucleus.
negative regulation of protein import into nucleus
Any process that stops, prevents or reduces the frequency, rate or extent of the movement of proteins from the cytoplasm into the nucleus.
regulation of transcription factor import into nucleus
Any process that modulates the frequency, rate or extent of the movement of a transcription factor from the cytoplasm to the nucleus.
transcription factor import into nucleus
The directed movement of a transcription factor from the cytoplasm to the nucleus.
negative regulation of transcription factor import into nucleus
Any process that stops, prevents or reduces the frequency, rate or extent of the movement of a transcription factor from the cytoplasm to the nucleus.
establishment of protein localization
The directed movement of a protein to a specific location.
regulation of nucleocytoplasmic transport
Any process that modulates the frequency, rate or extent of the directed movement of substances between the nucleus and the cytoplasm.
negative regulation of nucleocytoplasmic transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances between the cytoplasm and the nucleus.
intracellular transport
The directed movement of substances within a cell.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of transport
Any process that modulates the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
negative regulation of transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
nuclear transport
The directed movement of substances into, out of, or within the nucleus.
nuclear import
The directed movement of substances into the nucleus.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
regulation of protein transport
Any process that modulates the frequency, rate or extent of the directed movement of a protein into, out of, within or between cells.
negative regulation of protein transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of a protein into, out of, within or between cells.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular localization
Any process by which a substance or cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in, a specific location within or in the membrane of a cell.
establishment of localization in cell
The directed movement of a substance or cellular entity, such as a protein complex or organelle, to a specific location within, or in the membrane of, a cell.
regulation of cellular localization
Any process that modulates the frequency, rate or extent of a process by which a cell, a substance, or a cellular entity is transported to, or maintained in a specific location within or in the membrane of a cell.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of establishment of protein localization
Any process that modulates the frequency, rate or extent of the directed movement of a protein to a specific location.
all
This term is the most general term possible
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of localization
Any process that modulates the frequency, rate or extent of any process by which a cell, a substance, or a cellular entity is transported to, or maintained in, a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular localization
Any process by which a substance or cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in, a specific location within or in the membrane of a cell.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
establishment of localization in cell
The directed movement of a substance or cellular entity, such as a protein complex or organelle, to a specific location within, or in the membrane of, a cell.
regulation of cellular localization
Any process that modulates the frequency, rate or extent of a process by which a cell, a substance, or a cellular entity is transported to, or maintained in a specific location within or in the membrane of a cell.
regulation of cellular localization
Any process that modulates the frequency, rate or extent of a process by which a cell, a substance, or a cellular entity is transported to, or maintained in a specific location within or in the membrane of a cell.
regulation of transport
Any process that modulates the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
negative regulation of transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
protein transport
The directed movement of proteins into, out of, within or between cells.
intracellular transport
The directed movement of substances within a cell.
regulation of establishment of protein localization
Any process that modulates the frequency, rate or extent of the directed movement of a protein to a specific location.
regulation of intracellular transport
Any process that modulates the frequency, rate or extent of the directed movement of substances within cells.
negative regulation of transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
regulation of protein localization
Any process that modulates the frequency, rate or extent of any process by which a protein is transported to, or maintained in, a specific location.
cellular protein localization
Any process by which a protein is transported to, and/or maintained in, a specific location within or at the surface of a cell.
establishment of protein localization
The directed movement of a protein to a specific location.
intracellular protein transport
The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell.
regulation of protein transport
Any process that modulates the frequency, rate or extent of the directed movement of a protein into, out of, within or between cells.
negative regulation of protein transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of a protein into, out of, within or between cells.
intracellular protein transport
The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell.
regulation of intracellular transport
Any process that modulates the frequency, rate or extent of the directed movement of substances within cells.
negative regulation of intracellular transport
Any process that stops, prevents, or reduces the frequency, rate or extent of the directed movement of substances within cells.
regulation of protein transport
Any process that modulates the frequency, rate or extent of the directed movement of a protein into, out of, within or between cells.
negative regulation of intracellular transport
Any process that stops, prevents, or reduces the frequency, rate or extent of the directed movement of substances within cells.
regulation of intracellular protein transport
Any process that modulates the frequency, rate or extent of the directed movement of proteins within cells.
negative regulation of protein import into nucleus
Any process that stops, prevents or reduces the frequency, rate or extent of the movement of proteins from the cytoplasm into the nucleus.
regulation of intracellular protein transport
Any process that modulates the frequency, rate or extent of the directed movement of proteins within cells.
negative regulation of protein transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of a protein into, out of, within or between cells.
regulation of protein import into nucleus
Any process that modulates the frequency, rate or extent of movement of proteins from the cytoplasm to the nucleus.
negative regulation of nucleocytoplasmic transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances between the cytoplasm and the nucleus.
protein import into nucleus
The directed movement of a protein from the cytoplasm to the nucleus.
regulation of protein import into nucleus
Any process that modulates the frequency, rate or extent of movement of proteins from the cytoplasm to the nucleus.
negative regulation of protein import into nucleus
Any process that stops, prevents or reduces the frequency, rate or extent of the movement of proteins from the cytoplasm into the nucleus.
regulation of nucleocytoplasmic transport
Any process that modulates the frequency, rate or extent of the directed movement of substances between the nucleus and the cytoplasm.
negative regulation of nucleocytoplasmic transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances between the cytoplasm and the nucleus.
negative regulation of protein import into nucleus
Any process that stops, prevents or reduces the frequency, rate or extent of the movement of proteins from the cytoplasm into the nucleus.
regulation of transcription factor import into nucleus
Any process that modulates the frequency, rate or extent of the movement of a transcription factor from the cytoplasm to the nucleus.
negative regulation of transcription factor import into nucleus
Any process that stops, prevents or reduces the frequency, rate or extent of the movement of a transcription factor from the cytoplasm to the nucleus.
protein import into nucleus
The directed movement of a protein from the cytoplasm to the nucleus.
negative regulation of transcription factor import into nucleus
Any process that stops, prevents or reduces the frequency, rate or extent of the movement of a transcription factor from the cytoplasm to the nucleus.


AAASachalasia, adrenocortical insufficiency, alacrimia (Allgrove, triple-A) (218075_at), score: 0.59 ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.61 ACSL1acyl-CoA synthetase long-chain family member 1 (201963_at), score: -0.6 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.78 ACSS3acyl-CoA synthetase short-chain family member 3 (219616_at), score: 0.64 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.83 ADARadenosine deaminase, RNA-specific (201786_s_at), score: 0.67 ADORA2Badenosine A2b receptor (205891_at), score: 0.7 AHI1Abelson helper integration site 1 (221569_at), score: -0.72 AIM1absent in melanoma 1 (212543_at), score: 0.77 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.61 ANXA10annexin A10 (210143_at), score: -0.61 APPamyloid beta (A4) precursor protein (200602_at), score: -0.66 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (202211_at), score: -0.59 ARL6IP1ADP-ribosylation factor-like 6 interacting protein 1 (211935_at), score: 0.59 ATF5activating transcription factor 5 (204999_s_at), score: 0.54 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.65 BAT1HLA-B associated transcript 1 (200041_s_at), score: 0.57 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.61 BCL2L1BCL2-like 1 (215037_s_at), score: 0.55 BMP6bone morphogenetic protein 6 (206176_at), score: -0.83 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.55 C10orf97chromosome 10 open reading frame 97 (218297_at), score: -0.58 C11orf75chromosome 11 open reading frame 75 (219806_s_at), score: -0.66 C12orf29chromosome 12 open reading frame 29 (213701_at), score: -0.57 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.75 C14orf94chromosome 14 open reading frame 94 (218383_at), score: 0.73 C15orf33chromosome 15 open reading frame 33 (216411_s_at), score: -0.59 C16orf45chromosome 16 open reading frame 45 (212736_at), score: 0.6 C1orf107chromosome 1 open reading frame 107 (214193_s_at), score: -0.56 C20orf117chromosome 20 open reading frame 117 (207711_at), score: 0.6 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: -0.58 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.82 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.72 C7orf64chromosome 7 open reading frame 64 (221596_s_at), score: -0.62 CABYRcalcium binding tyrosine-(Y)-phosphorylation regulated (219928_s_at), score: 0.59 CALB2calbindin 2 (205428_s_at), score: -0.85 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.57 CAPN5calpain 5 (205166_at), score: 0.61 CASC3cancer susceptibility candidate 3 (207842_s_at), score: 0.61 CCNFcyclin F (204826_at), score: 0.68 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.77 CDC25Ccell division cycle 25 homolog C (S. pombe) (205167_s_at), score: 0.6 CDC2L6cell division cycle 2-like 6 (CDK8-like) (212899_at), score: 0.7 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.54 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: 0.72 CDCA3cell division cycle associated 3 (221436_s_at), score: 0.65 CDH13cadherin 13, H-cadherin (heart) (204726_at), score: -0.61 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: -0.65 CDKN2Dcyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) (210240_s_at), score: 0.69 CDKN3cyclin-dependent kinase inhibitor 3 (209714_s_at), score: 0.59 CENPAcentromere protein A (204962_s_at), score: 0.55 CENPEcentromere protein E, 312kDa (205046_at), score: 0.6 CENPFcentromere protein F, 350/400ka (mitosin) (207828_s_at), score: 0.61 CEP110centrosomal protein 110kDa (205642_at), score: 0.55 CHEK2CHK2 checkpoint homolog (S. pombe) (210416_s_at), score: 0.56 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.64 CICcapicua homolog (Drosophila) (212784_at), score: 0.71 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: 0.67 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.61 CLN5ceroid-lipofuscinosis, neuronal 5 (204084_s_at), score: -0.56 CMPK1cytidine monophosphate (UMP-CMP) kinase 1, cytosolic (217870_s_at), score: -0.58 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: 0.55 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.61 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.83 CScitrate synthase (208660_at), score: 0.66 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: -0.66 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.6 DDB2damage-specific DNA binding protein 2, 48kDa (203409_at), score: 0.54 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.58 DENND3DENN/MADD domain containing 3 (212974_at), score: 0.57 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.8 DIS3DIS3 mitotic control homolog (S. cerevisiae) (218362_s_at), score: -0.69 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: -0.64 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.92 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: -0.73 DOCK9dedicator of cytokinesis 9 (212538_at), score: -0.61 DOPEY1dopey family member 1 (40612_at), score: -0.68 DUSP4dual specificity phosphatase 4 (204014_at), score: -0.57 EAF2ELL associated factor 2 (219551_at), score: -0.74 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: -0.7 EDNRAendothelin receptor type A (204464_s_at), score: -0.59 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: -0.76 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: -0.66 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.7 ELOVL4elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 (219532_at), score: -0.56 EMCNendomucin (219436_s_at), score: 0.61 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.58 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.64 EVI1ecotropic viral integration site 1 (221884_at), score: -0.6 FABP3fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) (214285_at), score: -0.58 FAM155Afamily with sequence similarity 155, member A (214825_at), score: -0.58 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.72 FAM64Afamily with sequence similarity 64, member A (221591_s_at), score: 0.62 FAM8A1family with sequence similarity 8, member A1 (203420_at), score: -0.57 FAT4FAT tumor suppressor homolog 4 (Drosophila) (219427_at), score: -0.68 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.75 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: -0.56 FGF7fibroblast growth factor 7 (keratinocyte growth factor) (205782_at), score: -0.62 FICDFIC domain containing (219910_at), score: -0.65 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.58 FLOT1flotillin 1 (210142_x_at), score: 0.55 FOSL1FOS-like antigen 1 (204420_at), score: 0.58 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.68 FOXK2forkhead box K2 (203064_s_at), score: 0.57 FOXM1forkhead box M1 (202580_x_at), score: 0.62 FSTL3follistatin-like 3 (secreted glycoprotein) (203592_s_at), score: -0.56 FUBP3far upstream element (FUSE) binding protein 3 (212824_at), score: -0.57 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: -0.6 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.92 GHRgrowth hormone receptor (205498_at), score: -0.8 GKglycerol kinase (207387_s_at), score: -0.7 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.91 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.74 GMPRguanosine monophosphate reductase (204187_at), score: 0.67 GNL3Lguanine nucleotide binding protein-like 3 (nucleolar)-like (205010_at), score: -0.59 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.6 GPSM2G-protein signaling modulator 2 (AGS3-like, C. elegans) (221922_at), score: 0.63 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.58 GTSE1G-2 and S-phase expressed 1 (204318_s_at), score: 0.59 H1F0H1 histone family, member 0 (208886_at), score: 0.63 H1FXH1 histone family, member X (204805_s_at), score: 0.75 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: 0.7 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: -0.59 HIST1H1Chistone cluster 1, H1c (209398_at), score: 0.59 HJURPHolliday junction recognition protein (218726_at), score: 0.57 HMG20Bhigh-mobility group 20B (210719_s_at), score: 0.59 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.63 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: 0.58 HOXC5homeobox C5 (206739_at), score: -0.6 HRH1histamine receptor H1 (205580_s_at), score: 0.59 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.72 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.6 IL33interleukin 33 (209821_at), score: -0.56 IL9Rinterleukin 9 receptor (217212_s_at), score: -0.56 INSIG2insulin induced gene 2 (209566_at), score: -0.59 IRF1interferon regulatory factor 1 (202531_at), score: 0.55 IRS1insulin receptor substrate 1 (204686_at), score: -0.59 ISOC1isochorismatase domain containing 1 (218170_at), score: -0.57 ITGA1integrin, alpha 1 (214660_at), score: -0.63 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: -0.58 JUPjunction plakoglobin (201015_s_at), score: 0.67 KAL1Kallmann syndrome 1 sequence (205206_at), score: -1 KIAA0562KIAA0562 (204075_s_at), score: -0.67 KIAA1462KIAA1462 (213316_at), score: 0.62 KIF15kinesin family member 15 (219306_at), score: 0.58 KIF20Akinesin family member 20A (218755_at), score: 0.56 KLHL21kelch-like 21 (Drosophila) (203068_at), score: -0.61 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.62 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.74 LARSleucyl-tRNA synthetase (217810_x_at), score: -0.59 LIG1ligase I, DNA, ATP-dependent (202726_at), score: 0.58 LIN7Blin-7 homolog B (C. elegans) (219760_at), score: -0.63 LMNB2lamin B2 (216952_s_at), score: 0.55 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.63 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.59 LOC391132similar to hCG2041276 (216177_at), score: -0.75 LOC399491LOC399491 protein (214035_x_at), score: 0.58 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: -0.61 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.65 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.62 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.62 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.61 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.61 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: 0.57 MFAP3microfibrillar-associated protein 3 (213123_at), score: -0.59 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.8 MKI67antigen identified by monoclonal antibody Ki-67 (212022_s_at), score: 0.58 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.6 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: 0.6 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: -0.61 MYBBP1AMYB binding protein (P160) 1a (219098_at), score: -0.58 NARG1LNMDA receptor regulated 1-like (219378_at), score: -0.57 NCALDneurocalcin delta (211685_s_at), score: -0.74 NCAPD2non-SMC condensin I complex, subunit D2 (201774_s_at), score: 0.66 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.65 NEFLneurofilament, light polypeptide (221805_at), score: -0.6 NEFMneurofilament, medium polypeptide (205113_at), score: -0.86 NEK2NIMA (never in mitosis gene a)-related kinase 2 (204641_at), score: 0.55 NF1neurofibromin 1 (211094_s_at), score: 0.61 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.56 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.58 NMIN-myc (and STAT) interactor (203964_at), score: 0.59 NOX4NADPH oxidase 4 (219773_at), score: -0.86 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.71 NPTX1neuronal pentraxin I (204684_at), score: -0.57 NR5A2nuclear receptor subfamily 5, group A, member 2 (208343_s_at), score: -0.59 NRXN3neurexin 3 (205795_at), score: -0.61 NUDT1nudix (nucleoside diphosphate linked moiety X)-type motif 1 (204766_s_at), score: 0.55 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.56 OBFC2Aoligonucleotide/oligosaccharide-binding fold containing 2A (219334_s_at), score: -0.59 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.86 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.72 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.66 OR7E156Polfactory receptor, family 7, subfamily E, member 156 pseudogene (222327_x_at), score: -0.59 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.69 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.62 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.72 PCDH9protocadherin 9 (219737_s_at), score: -0.93 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.56 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.71 PIONpigeon homolog (Drosophila) (222150_s_at), score: -0.68 PITX1paired-like homeodomain 1 (208502_s_at), score: 0.58 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.65 PLK1polo-like kinase 1 (Drosophila) (202240_at), score: 0.7 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: 0.56 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.84 PTHLHparathyroid hormone-like hormone (211756_at), score: -0.64 PTTG1pituitary tumor-transforming 1 (203554_x_at), score: 0.56 PTTG3pituitary tumor-transforming 3 (208511_at), score: 0.6 PVRL3poliovirus receptor-related 3 (213325_at), score: -0.56 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.68 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: -0.66 RAB6CRAB6C, member RAS oncogene family (210406_s_at), score: -0.59 RAB7ARAB7A, member RAS oncogene family (211960_s_at), score: -0.56 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: 0.65 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.71 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.68 RNF11ring finger protein 11 (208924_at), score: -0.67 RNF220ring finger protein 220 (219988_s_at), score: 0.62 RNF44ring finger protein 44 (203286_at), score: 0.59 RP4-691N24.1ninein-like (207705_s_at), score: 0.58 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: -0.6 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.68 RPL39Lribosomal protein L39-like (210115_at), score: 0.57 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: -0.56 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.64 SAP30LSAP30-like (219129_s_at), score: 0.79 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.59 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.59 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.68 SDF2L1stromal cell-derived factor 2-like 1 (218681_s_at), score: -0.56 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: 0.69 SEPHS1selenophosphate synthetase 1 (208939_at), score: 0.65 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.65 SF1splicing factor 1 (208313_s_at), score: 0.67 SHROOM2shroom family member 2 (204967_at), score: -0.57 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.63 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: -0.63 SIX1SIX homeobox 1 (205817_at), score: 0.54 SLC11A2solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 (203124_s_at), score: -0.57 SLC25A32solute carrier family 25, member 32 (221020_s_at), score: -0.68 SLC2A4RGSLC2A4 regulator (218494_s_at), score: 0.54 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: -0.81 SLC39A14solute carrier family 39 (zinc transporter), member 14 (212110_at), score: -0.66 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: 0.67 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: -0.61 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.87 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: -0.69 SMAD3SMAD family member 3 (218284_at), score: 0.76 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.57 SMN1survival of motor neuron 1, telomeric (203852_s_at), score: -0.57 SNNstannin (218032_at), score: 0.66 SORBS1sorbin and SH3 domain containing 1 (218087_s_at), score: -0.59 SPAG5sperm associated antigen 5 (203145_at), score: 0.54 SSTR1somatostatin receptor 1 (208482_at), score: -0.69 STIM1stromal interaction molecule 1 (202764_at), score: 0.61 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.64 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: -0.74 SYNE2spectrin repeat containing, nuclear envelope 2 (202761_s_at), score: 0.64 SYNJ1synaptojanin 1 (212990_at), score: -0.61 TACC3transforming, acidic coiled-coil containing protein 3 (218308_at), score: 0.57 TAF13TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa (205966_at), score: -0.67 TAF2TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa (209523_at), score: -0.59 TASP1taspase, threonine aspartase, 1 (219443_at), score: -0.59 TBC1D8BTBC1 domain family, member 8B (with GRAM domain) (219771_at), score: -0.56 TDGthymine-DNA glycosylase (203743_s_at), score: -0.59 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.54 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.64 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.74 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.67 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.62 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.72 TLE4transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) (204872_at), score: -0.73 TMEM104transmembrane protein 104 (220097_s_at), score: -0.63 TMEM2transmembrane protein 2 (218113_at), score: -0.68 TMSB15Bthymosin beta 15B (214051_at), score: 0.6 TOMM20translocase of outer mitochondrial membrane 20 homolog (yeast) (200662_s_at), score: -0.68 TP53tumor protein p53 (201746_at), score: 0.62 TPBGtrophoblast glycoprotein (203476_at), score: 0.59 TPX2TPX2, microtubule-associated, homolog (Xenopus laevis) (210052_s_at), score: 0.55 TRAF5TNF receptor-associated factor 5 (204352_at), score: -0.58 TRAFD1TRAF-type zinc finger domain containing 1 (35254_at), score: 0.62 TRAPPC2trafficking protein particle complex 2 (219351_at), score: -0.6 TRIM21tripartite motif-containing 21 (204804_at), score: 0.58 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: -0.6 TROAPtrophinin associated protein (tastin) (204649_at), score: 0.59 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: -0.64 TRPC6transient receptor potential cation channel, subfamily C, member 6 (217287_s_at), score: -0.64 UBE2Wubiquitin-conjugating enzyme E2W (putative) (218521_s_at), score: -0.62 UFM1ubiquitin-fold modifier 1 (218050_at), score: -0.6 UGDHUDP-glucose dehydrogenase (203343_at), score: -0.64 URB1URB1 ribosome biogenesis 1 homolog (S. cerevisiae) (212996_s_at), score: -0.76 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: -0.6 WDR4WD repeat domain 4 (221632_s_at), score: -0.6 WDR42AWD repeat domain 42A (202249_s_at), score: 0.59 WDR6WD repeat domain 6 (217734_s_at), score: 0.66 WDR62WD repeat domain 62 (215218_s_at), score: 0.58 WHAMML1WAS protein homolog associated with actin, golgi membranes and microtubules-like 1 (213908_at), score: -0.56 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.66 YTHDC2YTH domain containing 2 (213077_at), score: -0.63 ZBTB20zinc finger and BTB domain containing 20 (205383_s_at), score: 0.59 ZBTB6zinc finger and BTB domain containing 6 (206098_at), score: -0.68 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: -0.63 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.8 ZFAND1zinc finger, AN1-type domain 1 (218919_at), score: -0.6 ZNF20zinc finger protein 20 (213916_at), score: 0.59 ZNF23zinc finger protein 23 (KOX 16) (213934_s_at), score: -0.74 ZNF395zinc finger protein 395 (218149_s_at), score: 0.55 ZNF652zinc finger protein 652 (205594_at), score: -0.61 ZNF706zinc finger protein 706 (218059_at), score: 0.55
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |