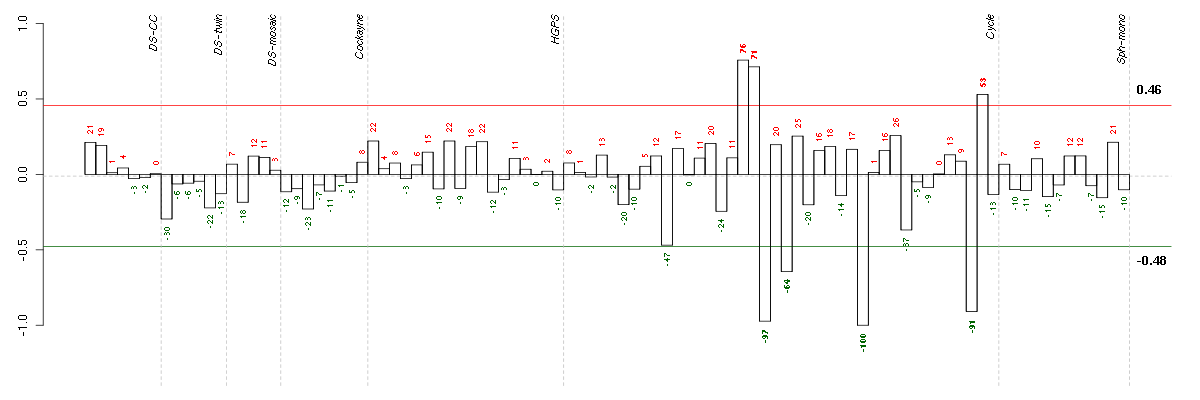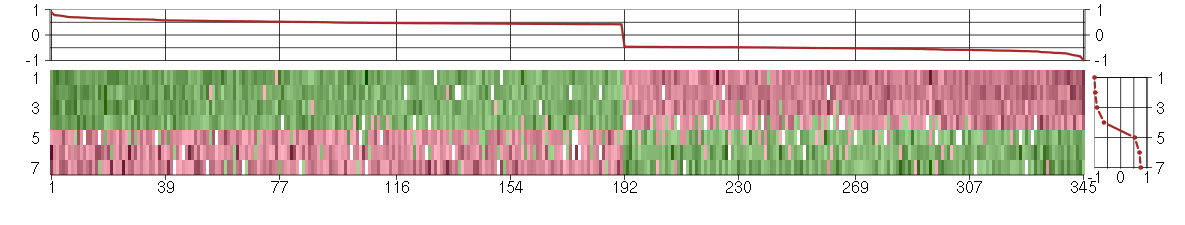

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
receptor binding
Interacting selectively with one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function.
hormone activity
The action characteristic of a hormone, any substance formed in very small amounts in one specialized organ or group of cells and carried (sometimes in the bloodstream) to another organ or group of cells in the same organism, upon which it has a specific regulatory action. The term was originally applied to agents with a stimulatory physiological action in vertebrate animals (as opposed to a chalone, which has a depressant action). Usage is now extended to regulatory compounds in lower animals and plants, and to synthetic substances having comparable effects; all bind receptors and trigger some biological process.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
all
This term is the most general term possible
ABL1c-abl oncogene 1, receptor tyrosine kinase (202123_s_at), score: 0.46 ACANaggrecan (205679_x_at), score: -0.53 ADAM21ADAM metallopeptidase domain 21 (207665_at), score: -0.57 ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (220866_at), score: 0.62 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.54 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: -0.52 AGRNagrin (212285_s_at), score: 0.43 AJAP1adherens junctions associated protein 1 (206460_at), score: 0.42 ANKRD28ankyrin repeat domain 28 (213035_at), score: -0.68 ANXA10annexin A10 (210143_at), score: 0.63 AOF2amine oxidase (flavin containing) domain 2 (212348_s_at), score: -0.5 AP3S2adaptor-related protein complex 3, sigma 2 subunit (202399_s_at), score: -0.48 ARHGEF11Rho guanine nucleotide exchange factor (GEF) 11 (202914_s_at), score: 0.52 ARL17ADP-ribosylation factor-like 17 (210435_at), score: 0.48 ARNT2aryl-hydrocarbon receptor nuclear translocator 2 (202986_at), score: -0.59 ASTN2astrotactin 2 (209693_at), score: -0.61 ATXN2ataxin 2 (202622_s_at), score: -0.5 B3GNT2UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (219326_s_at), score: 0.51 B3GNTL1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 (213589_s_at), score: -0.77 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (221484_at), score: 0.54 BAHD1bromo adjacent homology domain containing 1 (203051_at), score: 0.46 BAXBCL2-associated X protein (208478_s_at), score: -0.62 BCCIPBRCA2 and CDKN1A interacting protein (218264_at), score: -0.62 BRIP1BRCA1 interacting protein C-terminal helicase 1 (221703_at), score: 0.47 C11orf67chromosome 11 open reading frame 67 (221600_s_at), score: -0.61 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: 0.45 C16orf7chromosome 16 open reading frame 7 (205781_at), score: 0.5 C17orf39chromosome 17 open reading frame 39 (220058_at), score: -0.53 C19orf22chromosome 19 open reading frame 22 (221764_at), score: 0.5 C19orf54chromosome 19 open reading frame 54 (222052_at), score: -0.58 C1orf159chromosome 1 open reading frame 159 (219337_at), score: 0.51 C1orf50chromosome 1 open reading frame 50 (219406_at), score: -0.5 C1orf9chromosome 1 open reading frame 9 (203429_s_at), score: 0.48 C2CD2LC2CD2-like (204757_s_at), score: 0.5 C5orf4chromosome 5 open reading frame 4 (48031_r_at), score: -0.48 C6orf62chromosome 6 open reading frame 62 (213875_x_at), score: -0.51 CA12carbonic anhydrase XII (203963_at), score: -0.49 CAMK2Bcalcium/calmodulin-dependent protein kinase II beta (34846_at), score: 0.46 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: 0.43 CCNE1cyclin E1 (213523_at), score: 0.57 CD320CD320 molecule (218529_at), score: -0.7 CD40CD40 molecule, TNF receptor superfamily member 5 (35150_at), score: 0.44 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: 0.53 CDC25Acell division cycle 25 homolog A (S. pombe) (204695_at), score: 0.51 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.46 CDCA4cell division cycle associated 4 (218399_s_at), score: 0.66 CDCP1CUB domain containing protein 1 (218451_at), score: 0.51 CDH18cadherin 18, type 2 (206280_at), score: -0.63 CDKL3cyclin-dependent kinase-like 3 (219831_at), score: -0.53 CDT1chromatin licensing and DNA replication factor 1 (209832_s_at), score: 0.47 CENPTcentromere protein T (218148_at), score: 0.46 CEP70centrosomal protein 70kDa (219036_at), score: -0.53 CHRM2cholinergic receptor, muscarinic 2 (221330_at), score: -0.47 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: 0.89 CLEC1AC-type lectin domain family 1, member A (219761_at), score: 0.45 CLEC3BC-type lectin domain family 3, member B (205200_at), score: 0.7 CLUclusterin (208791_at), score: 0.54 CNNM2cyclin M2 (206818_s_at), score: -0.55 CNNM3cyclin M3 (220739_s_at), score: 0.54 COL14A1collagen, type XIV, alpha 1 (212865_s_at), score: 0.45 COL5A3collagen, type V, alpha 3 (52255_s_at), score: 0.78 COL7A1collagen, type VII, alpha 1 (217312_s_at), score: 0.67 COQ3coenzyme Q3 homolog, methyltransferase (S. cerevisiae) (221227_x_at), score: -0.48 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: 0.54 CRYL1crystallin, lambda 1 (220753_s_at), score: 0.46 CST6cystatin E/M (206595_at), score: 0.57 CTNScystinosis, nephropathic (36566_at), score: -0.47 CTSKcathepsin K (202450_s_at), score: 0.48 CTSOcathepsin O (203758_at), score: 0.55 CTSZcathepsin Z (210042_s_at), score: -0.5 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: 0.6 CYP2A6cytochrome P450, family 2, subfamily A, polypeptide 6 (211295_x_at), score: 0.47 CYTH1cytohesin 1 (202880_s_at), score: 0.56 DARS2aspartyl-tRNA synthetase 2, mitochondrial (218365_s_at), score: -0.48 DDIT3DNA-damage-inducible transcript 3 (209383_at), score: -0.46 DEM1defects in morphology 1 homolog (S. cerevisiae) (219567_s_at), score: 0.48 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: 0.7 DNAJC17DnaJ (Hsp40) homolog, subfamily C, member 17 (219861_at), score: -0.47 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (213853_at), score: -0.54 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (202416_at), score: -0.49 DNM3dynamin 3 (209839_at), score: 0.48 DPH5DPH5 homolog (S. cerevisiae) (219590_x_at), score: -0.54 DPP4dipeptidyl-peptidase 4 (211478_s_at), score: 0.52 DUS4Ldihydrouridine synthase 4-like (S. cerevisiae) (205761_s_at), score: -0.48 DUSP5dual specificity phosphatase 5 (209457_at), score: 0.65 DUSP6dual specificity phosphatase 6 (208891_at), score: 0.76 EDN1endothelin 1 (218995_s_at), score: 0.54 EDNRBendothelin receptor type B (204271_s_at), score: 0.64 EGFL6EGF-like-domain, multiple 6 (219454_at), score: 0.45 EGR2early growth response 2 (Krox-20 homolog, Drosophila) (205249_at), score: 0.51 EGR3early growth response 3 (206115_at), score: 0.58 EIF3Heukaryotic translation initiation factor 3, subunit H (201592_at), score: -0.48 EIF4A2eukaryotic translation initiation factor 4A, isoform 2 (200912_s_at), score: -0.47 EIF4EBP2eukaryotic translation initiation factor 4E binding protein 2 (208770_s_at), score: 0.52 ELA3Aelastase 3A, pancreatic (211738_x_at), score: 0.42 ELNelastin (212670_at), score: 0.44 EMCNendomucin (219436_s_at), score: -0.81 EML1echinoderm microtubule associated protein like 1 (204797_s_at), score: 0.52 EPOerythropoietin (217254_s_at), score: -0.48 EREGepiregulin (205767_at), score: 0.53 ERO1LERO1-like (S. cerevisiae) (218498_s_at), score: 0.46 ESRRAestrogen-related receptor alpha (1487_at), score: 0.56 EVI2Aecotropic viral integration site 2A (204774_at), score: -0.53 EVI2Becotropic viral integration site 2B (211742_s_at), score: -0.64 EVLEnah/Vasp-like (217838_s_at), score: 0.47 EXOSC5exosome component 5 (218481_at), score: -0.55 FAM117Afamily with sequence similarity 117, member A (221249_s_at), score: -0.58 FAM168Bfamily with sequence similarity 168, member B (212017_at), score: 0.52 FAM171A1family with sequence similarity 171, member A1 (212771_at), score: -0.51 FAM29Afamily with sequence similarity 29, member A (218602_s_at), score: 0.52 FAM48Afamily with sequence similarity 48, member A (221774_x_at), score: -0.58 FAM63Bfamily with sequence similarity 63, member B (214691_x_at), score: 0.56 FBXO31F-box protein 31 (219785_s_at), score: -0.63 FEM1Bfem-1 homolog b (C. elegans) (212367_at), score: 0.47 FEM1Cfem-1 homolog c (C. elegans) (213341_at), score: 0.54 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.61 FOXK2forkhead box K2 (203064_s_at), score: 0.45 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: 0.51 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: -0.72 GAAglucosidase, alpha; acid (202812_at), score: 0.68 GALNSgalactosamine (N-acetyl)-6-sulfate sulfatase (206335_at), score: -0.54 GALTgalactose-1-phosphate uridylyltransferase (203179_at), score: -0.48 GANgigaxonin (220124_at), score: 0.48 GIT1G protein-coupled receptor kinase interacting ArfGAP 1 (218030_at), score: 0.46 GKglycerol kinase (207387_s_at), score: 0.51 GLTSCR2glioma tumor suppressor candidate region gene 2 (217807_s_at), score: -0.49 GNB1Lguanine nucleotide binding protein (G protein), beta polypeptide 1-like (220762_s_at), score: -0.56 GNPDA1glucosamine-6-phosphate deaminase 1 (202382_s_at), score: -0.48 GPM6Bglycoprotein M6B (209167_at), score: 0.68 GPR137G protein-coupled receptor 137 (43934_at), score: 0.44 GPR153G protein-coupled receptor 153 (221902_at), score: 0.52 GPR177G protein-coupled receptor 177 (221958_s_at), score: 0.46 GPR183G protein-coupled receptor 183 (205419_at), score: 0.46 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: -0.6 GPSM3G-protein signaling modulator 3 (AGS3-like, C. elegans) (204265_s_at), score: -0.47 HBEGFheparin-binding EGF-like growth factor (203821_at), score: 0.47 HERC3hect domain and RLD 3 (206183_s_at), score: 0.43 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 0.63 HINFPhistone H4 transcription factor (206495_s_at), score: -0.54 HISPPD2Ahistidine acid phosphatase domain containing 2A (204578_at), score: -0.47 HIST1H2AGhistone cluster 1, H2ag (207156_at), score: -0.52 HIST1H2BFhistone cluster 1, H2bf (208490_x_at), score: 0.44 HIST1H2BKhistone cluster 1, H2bk (209806_at), score: 0.49 HIST1H4Hhistone cluster 1, H4h (208180_s_at), score: 0.55 HLXH2.0-like homeobox (214438_at), score: 0.42 HOXD1homeobox D1 (205975_s_at), score: 0.42 HSPB2heat shock 27kDa protein 2 (205824_at), score: -0.58 HSPBAP1HSPB (heat shock 27kDa) associated protein 1 (219284_at), score: -0.57 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.46 IER2immediate early response 2 (202081_at), score: 0.56 IFRD1interferon-related developmental regulator 1 (202147_s_at), score: -0.46 IGF2insulin-like growth factor 2 (somatomedin A) (202409_at), score: 0.54 IGF2BP2insulin-like growth factor 2 mRNA binding protein 2 (218847_at), score: -0.46 IL11interleukin 11 (206924_at), score: 0.43 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: 0.61 IL33interleukin 33 (209821_at), score: 0.77 INHAinhibin, alpha (210141_s_at), score: -0.47 INTS8integrator complex subunit 8 (218905_at), score: -0.53 IPPKinositol 1,3,4,5,6-pentakisphosphate 2-kinase (219092_s_at), score: 0.63 JMJD1Ajumonji domain containing 1A (212689_s_at), score: -0.48 JUNBjun B proto-oncogene (201473_at), score: 0.62 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.43 KCNE4potassium voltage-gated channel, Isk-related family, member 4 (222379_at), score: -0.61 KCNJ15potassium inwardly-rectifying channel, subfamily J, member 15 (210119_at), score: 0.63 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: 0.45 KCTD14potassium channel tetramerisation domain containing 14 (58916_at), score: -0.49 KIAA0586KIAA0586 (205631_at), score: -0.58 KITv-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (205051_s_at), score: -0.61 KLHL5kelch-like 5 (Drosophila) (220682_s_at), score: -0.48 KRT17keratin 17 (212236_x_at), score: 0.51 KRT33Akeratin 33A (208483_x_at), score: 0.61 LEMD3LEM domain containing 3 (218604_at), score: 0.47 LIFleukemia inhibitory factor (cholinergic differentiation factor) (205266_at), score: 0.65 LMAN2Llectin, mannose-binding 2-like (221274_s_at), score: -0.6 LOC100128223hypothetical protein LOC100128223 (221264_s_at), score: -0.72 LOC388152hypothetical LOC388152 (220602_s_at), score: -0.68 LOC90379hypothetical protein BC002926 (221849_s_at), score: 0.43 LRP1low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) (200785_s_at), score: 0.45 LRP2low density lipoprotein-related protein 2 (205710_at), score: 0.46 LTBP3latent transforming growth factor beta binding protein 3 (219922_s_at), score: 0.46 MADCAM1mucosal vascular addressin cell adhesion molecule 1 (208037_s_at), score: 0.42 MAN1B1mannosidase, alpha, class 1B, member 1 (65884_at), score: 0.47 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.74 MAP7D3MAP7 domain containing 3 (219576_at), score: -0.51 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: 0.56 MED16mediator complex subunit 16 (43544_at), score: -0.48 MEF2Amyocyte enhancer factor 2A (214684_at), score: 0.46 MID1IP1MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) (218251_at), score: 0.57 MLPHmelanophilin (218211_s_at), score: -0.62 MMACHCmethylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria (211774_s_at), score: -0.49 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: 0.46 MSNmoesin (200600_at), score: 0.44 MTHFSDmethenyltetrahydrofolate synthetase domain containing (218879_s_at), score: -0.56 MTMR3myotubularin related protein 3 (202197_at), score: 0.43 MUSKmuscle, skeletal, receptor tyrosine kinase (207633_s_at), score: 0.57 N6AMT1N-6 adenine-specific DNA methyltransferase 1 (putative) (220311_at), score: -0.47 NAGAN-acetylgalactosaminidase, alpha- (202943_s_at), score: -0.5 NAGPAN-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (205090_s_at), score: -0.52 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.47 NEDD4Lneural precursor cell expressed, developmentally down-regulated 4-like (212445_s_at), score: 0.45 NEFLneurofilament, light polypeptide (221805_at), score: 0.59 NELFnasal embryonic LHRH factor (221214_s_at), score: 0.53 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: 0.72 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.44 NKX3-1NK3 homeobox 1 (209706_at), score: 0.43 NMBneuromedin B (205204_at), score: -0.52 NME6non-metastatic cells 6, protein expressed in (nucleoside-diphosphate kinase) (205851_at), score: -0.54 NOL8nucleolar protein 8 (218244_at), score: -0.59 NR2F1nuclear receptor subfamily 2, group F, member 1 (209505_at), score: 0.44 NR5A2nuclear receptor subfamily 5, group A, member 2 (208343_s_at), score: 0.44 NSUN5BNOL1/NOP2/Sun domain family, member 5B (214100_x_at), score: -0.47 NTHL1nth endonuclease III-like 1 (E. coli) (209731_at), score: -0.52 NUDT1nudix (nucleoside diphosphate linked moiety X)-type motif 1 (204766_s_at), score: -0.52 NUDT6nudix (nucleoside diphosphate linked moiety X)-type motif 6 (220183_s_at), score: -0.47 NUPL1nucleoporin like 1 (204435_at), score: 0.43 OLFML2Aolfactomedin-like 2A (213075_at), score: 0.56 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: 0.47 PAFAH1B3platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa (203228_at), score: -0.53 PAN2PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (203117_s_at), score: -0.47 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: -0.5 PENKproenkephalin (213791_at), score: 0.5 PER2period homolog 2 (Drosophila) (205251_at), score: 0.45 PFDN6prefoldin subunit 6 (222029_x_at), score: -0.49 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: -0.59 PKD1polycystic kidney disease 1 (autosomal dominant) (202328_s_at), score: 0.45 PKIAprotein kinase (cAMP-dependent, catalytic) inhibitor alpha (204612_at), score: 0.43 PLAURplasminogen activator, urokinase receptor (211924_s_at), score: 0.51 PLCXD1phosphatidylinositol-specific phospholipase C, X domain containing 1 (218951_s_at), score: 0.54 PLK3polo-like kinase 3 (Drosophila) (204958_at), score: 0.56 PLXNB1plexin B1 (215807_s_at), score: 0.57 PMAIP1phorbol-12-myristate-13-acetate-induced protein 1 (204285_s_at), score: 0.57 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.45 POP1processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) (213449_at), score: -0.47 PPFIBP1PTPRF interacting protein, binding protein 1 (liprin beta 1) (214374_s_at), score: -0.48 PPP2R1Bprotein phosphatase 2 (formerly 2A), regulatory subunit A, beta isoform (202884_s_at), score: 0.45 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: 0.65 PRSS3protease, serine, 3 (213421_x_at), score: 0.66 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: -0.6 PTCD1pentatricopeptide repeat domain 1 (218956_s_at), score: -0.74 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: 0.61 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: 0.52 PYCARDPYD and CARD domain containing (221666_s_at), score: -0.47 QSOX1quiescin Q6 sulfhydryl oxidase 1 (201482_at), score: 0.42 R3HDM2R3H domain containing 2 (203831_at), score: -0.52 RAB8BRAB8B, member RAS oncogene family (219210_s_at), score: -0.47 RABL3RAB, member of RAS oncogene family-like 3 (213970_at), score: -0.67 RALGDSral guanine nucleotide dissociation stimulator (209050_s_at), score: -0.7 RASA2RAS p21 protein activator 2 (206636_at), score: 0.63 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: 0.48 RBM4BRNA binding motif protein 4B (209497_s_at), score: -0.58 RCC1regulator of chromosome condensation 1 (206499_s_at), score: -0.52 RFX7regulatory factor X, 7 (218430_s_at), score: -0.58 RHBDD3rhomboid domain containing 3 (204402_at), score: 0.44 RHOBTB1Rho-related BTB domain containing 1 (212651_at), score: -0.48 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: 0.69 RNF44ring finger protein 44 (203286_at), score: -0.56 RPS6KA5ribosomal protein S6 kinase, 90kDa, polypeptide 5 (204633_s_at), score: 0.46 S100A3S100 calcium binding protein A3 (206027_at), score: -0.46 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: 0.63 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: 0.43 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: 0.43 SERPING1serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 (200986_at), score: 0.43 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: -0.85 SEZ6L2seizure related 6 homolog (mouse)-like 2 (218720_x_at), score: 0.48 SF1splicing factor 1 (208313_s_at), score: 0.47 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: -0.59 SLC16A6solute carrier family 16, member 6 (monocarboxylic acid transporter 7) (207038_at), score: 0.53 SLC17A7solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 (204230_s_at), score: 0.55 SLC20A2solute carrier family 20 (phosphate transporter), member 2 (202744_at), score: 0.58 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (205896_at), score: 0.48 SLC25A17solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 (211754_s_at), score: 0.47 SLC25A28solute carrier family 25, member 28 (221432_s_at), score: 0.56 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: -0.8 SLKSTE20-like kinase (yeast) (206875_s_at), score: 0.43 SMAD6SMAD family member 6 (207069_s_at), score: -0.51 SMU1smu-1 suppressor of mec-8 and unc-52 homolog (C. elegans) (218393_s_at), score: -0.51 SNHG3small nucleolar RNA host gene 3 (non-protein coding) (215011_at), score: -0.49 SNORA21small nucleolar RNA, H/ACA box 21 (215224_at), score: -0.62 SNX13sorting nexin 13 (213292_s_at), score: -0.51 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.47 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: 0.55 SPA17sperm autoantigenic protein 17 (205406_s_at), score: -0.56 SPAG1sperm associated antigen 1 (210117_at), score: 0.53 SPRY2sprouty homolog 2 (Drosophila) (204011_at), score: 0.54 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: 0.61 STC1stanniocalcin 1 (204597_x_at), score: 0.46 SYNMsynemin, intermediate filament protein (212730_at), score: -0.49 SYT1synaptotagmin I (203999_at), score: 0.49 SYT11synaptotagmin XI (209198_s_at), score: -0.7 TACC2transforming, acidic coiled-coil containing protein 2 (202289_s_at), score: -0.48 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.44 TAS2R9taste receptor, type 2, member 9 (221461_at), score: 0.42 TBC1D10BTBC1 domain family, member 10B (220947_s_at), score: 0.43 TBX2T-box 2 (40560_at), score: 0.45 TGFBRAP1transforming growth factor, beta receptor associated protein 1 (205210_at), score: -0.48 THOC2THO complex 2 (212994_at), score: -0.52 THOC6THO complex 6 homolog (Drosophila) (218848_at), score: -0.53 TIMM9translocase of inner mitochondrial membrane 9 homolog (yeast) (218316_at), score: -0.53 TLE2transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) (40837_at), score: 0.44 TMEM158transmembrane protein 158 (213338_at), score: 0.49 TMEM160transmembrane protein 160 (219219_at), score: -0.61 TMEM22transmembrane protein 22 (219569_s_at), score: 0.49 TMSB15Athymosin beta 15a (205347_s_at), score: -0.63 TNFAIP3tumor necrosis factor, alpha-induced protein 3 (202644_s_at), score: 0.45 TNIKTRAF2 and NCK interacting kinase (213107_at), score: -0.59 TOXthymocyte selection-associated high mobility group box (204529_s_at), score: -0.58 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: 0.69 TRAFD1TRAF-type zinc finger domain containing 1 (35254_at), score: 0.42 TRAPPC10trafficking protein particle complex 10 (209412_at), score: 0.43 TRIAP1TP53 regulated inhibitor of apoptosis 1 (218403_at), score: -0.65 TRIB1tribbles homolog 1 (Drosophila) (202241_at), score: 0.43 TRIP11thyroid hormone receptor interactor 11 (209778_at), score: -0.54 UBE3Aubiquitin protein ligase E3A (213291_s_at), score: 0.45 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: -1 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.45 USE1unconventional SNARE in the ER 1 homolog (S. cerevisiae) (221706_s_at), score: -0.55 USP36ubiquitin specific peptidase 36 (220370_s_at), score: 0.55 USP47ubiquitin specific peptidase 47 (221518_s_at), score: -0.48 VAMP1vesicle-associated membrane protein 1 (synaptobrevin 1) (213326_at), score: -0.64 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.51 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: -0.54 WDR3WD repeat domain 3 (218882_s_at), score: -0.48 WDR5BWD repeat domain 5B (219538_at), score: -0.47 XPO4exportin 4 (218479_s_at), score: -0.67 YEATS4YEATS domain containing 4 (218911_at), score: 0.42 YIPF1Yip1 domain family, member 1 (214733_s_at), score: -0.51 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.48 YY2YY2 transcription factor (216531_at), score: -0.51 ZBTB24zinc finger and BTB domain containing 24 (205340_at), score: -0.55 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: 0.62 ZFYVE9zinc finger, FYVE domain containing 9 (204893_s_at), score: -0.48 ZHX2zinc fingers and homeoboxes 2 (203556_at), score: -0.47 ZMAT5zinc finger, matrin type 5 (218752_at), score: -0.51 ZNF133zinc finger protein 133 (216960_s_at), score: -0.5 ZNF195zinc finger protein 195 (204234_s_at), score: -0.53 ZNF219zinc finger protein 219 (219314_s_at), score: 0.65 ZNF22zinc finger protein 22 (KOX 15) (218005_at), score: -0.63 ZNF239zinc finger protein 239 (206261_at), score: -0.48 ZNF248zinc finger protein 248 (213269_at), score: -0.46 ZNF3zinc finger protein 3 (212684_at), score: -0.48 ZNF337zinc finger protein 337 (37860_at), score: -0.5 ZNF443zinc finger protein 443 (205928_at), score: 0.44 ZNF480zinc finger protein 480 (222283_at), score: -0.48 ZNF667zinc finger protein 667 (207120_at), score: -0.53 ZRANB1zinc finger, RAN-binding domain containing 1 (201219_at), score: -0.51
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |