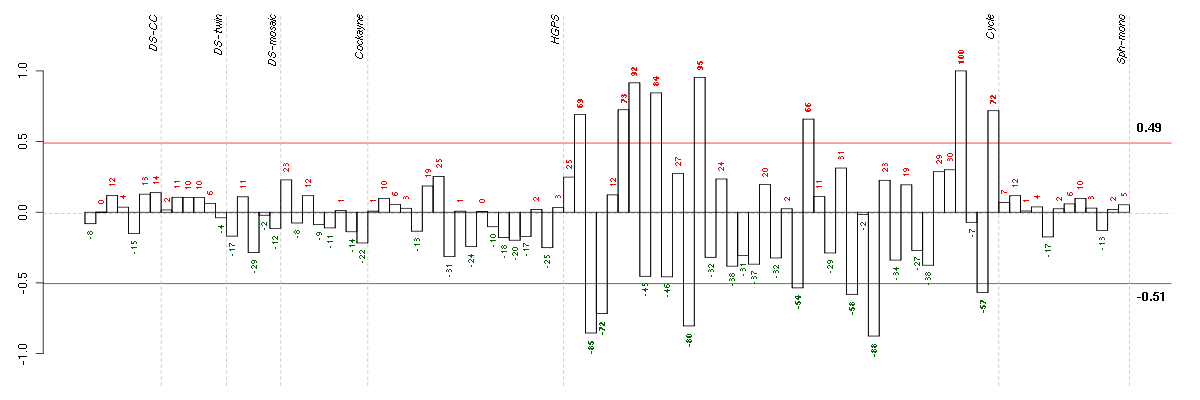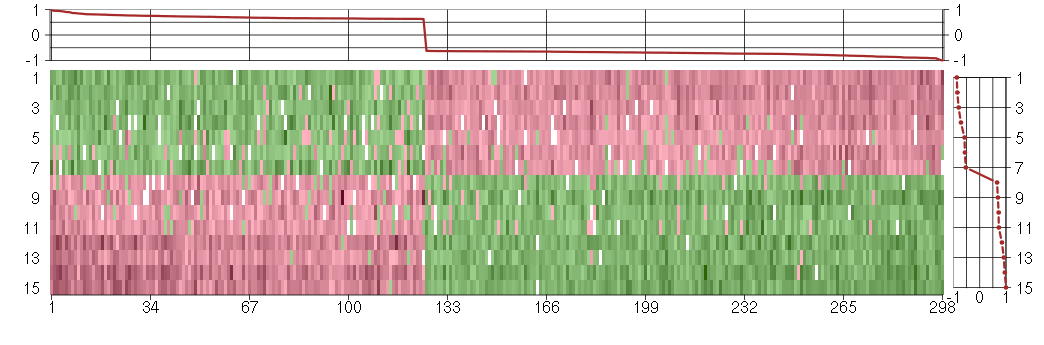

Under-expression is coded with green,
over-expression with red color.



AAASachalasia, adrenocortical insufficiency, alacrimia (Allgrove, triple-A) (218075_at), score: -0.66 ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: 0.65 ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: -0.71 ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.79 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: 0.69 ACTN3actinin, alpha 3 (206891_at), score: 0.66 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: 0.82 ADAadenosine deaminase (204639_at), score: 0.65 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.8 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: -0.7 AIM1absent in melanoma 1 (212543_at), score: -0.73 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: 0.71 APPamyloid beta (A4) precursor protein (200602_at), score: 0.76 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: -0.64 ASF1BASF1 anti-silencing function 1 homolog B (S. cerevisiae) (218115_at), score: -0.63 ATF5activating transcription factor 5 (204999_s_at), score: -0.79 ATMataxia telangiectasia mutated (210858_x_at), score: 0.71 ATN1atrophin 1 (40489_at), score: -0.66 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: 0.76 ATP8B1ATPase, class I, type 8B, member 1 (214594_x_at), score: 0.64 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: -0.67 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: -0.72 BAT2HLA-B associated transcript 2 (212081_x_at), score: -0.85 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: -0.64 BCL2L1BCL2-like 1 (215037_s_at), score: -0.73 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.82 C10orf97chromosome 10 open reading frame 97 (218297_at), score: 0.63 C11orf75chromosome 11 open reading frame 75 (219806_s_at), score: 0.65 C14orf45chromosome 14 open reading frame 45 (220173_at), score: 0.74 C14orf94chromosome 14 open reading frame 94 (218383_at), score: -0.73 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: -0.63 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: -0.75 C1orf38chromosome 1 open reading frame 38 (210785_s_at), score: -0.64 C20orf27chromosome 20 open reading frame 27 (50314_i_at), score: -0.65 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: 0.66 C4orf43chromosome 4 open reading frame 43 (218513_at), score: 0.77 C5orf44chromosome 5 open reading frame 44 (218674_at), score: 0.74 CABIN1calcineurin binding protein 1 (37652_at), score: -0.65 CALB2calbindin 2 (205428_s_at), score: 0.9 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: -0.72 CAPN5calpain 5 (205166_at), score: -0.71 CASC3cancer susceptibility candidate 3 (207842_s_at), score: -0.71 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: 0.7 CD248CD248 molecule, endosialin (219025_at), score: -0.67 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: -0.74 CDC2L6cell division cycle 2-like 6 (CDK8-like) (212899_at), score: -0.63 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.88 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: -0.65 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: -0.86 CDH13cadherin 13, H-cadherin (heart) (204726_at), score: 0.73 CDKN2Dcyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) (210240_s_at), score: -0.63 CICcapicua homolog (Drosophila) (212784_at), score: -0.89 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: -0.79 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: -0.78 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: -0.7 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.9 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.63 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: -0.67 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.89 CScitrate synthase (208660_at), score: -0.76 CUX1cut-like homeobox 1 (214743_at), score: 0.67 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: 0.65 CYP7B1cytochrome P450, family 7, subfamily B, polypeptide 1 (207386_at), score: -0.64 DDB2damage-specific DNA binding protein 2, 48kDa (203409_at), score: -0.65 DENND3DENN/MADD domain containing 3 (212974_at), score: -0.81 DENND4CDENN/MADD domain containing 4C (205684_s_at), score: 0.65 DENND5BDENN/MADD domain containing 5B (215058_at), score: 0.78 DLSTPdihydrolipoamide S-succinyltransferase pseudogene (E2 component of 2-oxo-glutarate complex) (215210_s_at), score: 0.65 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: -0.73 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: -0.73 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: 0.9 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (213853_at), score: 0.64 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: 0.76 DNM2dynamin 2 (202253_s_at), score: -0.63 DOPEY1dopey family member 1 (40612_at), score: 0.72 DSCR6Down syndrome critical region gene 6 (207267_s_at), score: -0.64 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.66 ECM1extracellular matrix protein 1 (209365_s_at), score: -0.65 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: 0.75 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: 0.69 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: -0.66 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: 0.63 EPHA4EPH receptor A4 (206114_at), score: 0.81 ERBB2IPerbb2 interacting protein (217941_s_at), score: 0.65 EVLEnah/Vasp-like (217838_s_at), score: -0.63 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: 0.81 FAT4FAT tumor suppressor homolog 4 (Drosophila) (219427_at), score: 0.75 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: -0.88 FBXO2F-box protein 2 (219305_x_at), score: 0.72 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: 0.66 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: 0.64 FHOD1formin homology 2 domain containing 1 (218530_at), score: -0.64 FKRPfukutin related protein (219853_at), score: -0.65 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.74 FLJ40113golgi autoantigen, golgin subfamily a-like pseudogene (213212_x_at), score: 0.66 FOSL1FOS-like antigen 1 (204420_at), score: -0.71 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: -0.76 FOXK2forkhead box K2 (203064_s_at), score: -0.92 FOXM1forkhead box M1 (202580_x_at), score: -0.66 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: 0.64 GABARAPL1GABA(A) receptor-associated protein like 1 (208868_s_at), score: 0.71 GABARAPL3GABA(A) receptors associated protein like 3 (pseudogene) (211458_s_at), score: 0.68 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: 0.77 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: -0.78 GHRgrowth hormone receptor (205498_at), score: 0.64 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: 0.8 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: 0.68 GMPRguanosine monophosphate reductase (204187_at), score: -0.65 GNL3Lguanine nucleotide binding protein-like 3 (nucleolar)-like (205010_at), score: 0.69 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: 0.66 H1F0H1 histone family, member 0 (208886_at), score: -0.75 H1FXH1 histone family, member X (204805_s_at), score: -0.86 HAB1B1 for mucin (215778_x_at), score: 0.8 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: -0.69 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.7 HIST1H1Chistone cluster 1, H1c (209398_at), score: -0.67 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: 0.66 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.89 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: 0.67 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: -0.68 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.92 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: -0.69 IDUAiduronidase, alpha-L- (205059_s_at), score: -0.76 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: 0.63 IL9Rinterleukin 9 receptor (217212_s_at), score: 0.7 INPP1inositol polyphosphate-1-phosphatase (202794_at), score: -0.65 INTS1integrator complex subunit 1 (212212_s_at), score: -0.63 ITGA1integrin, alpha 1 (214660_at), score: 0.78 JUNDjun D proto-oncogene (203751_x_at), score: -0.68 JUPjunction plakoglobin (201015_s_at), score: -0.86 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.95 KCTD15potassium channel tetramerisation domain containing 15 (218553_s_at), score: 0.66 KCTD2potassium channel tetramerisation domain containing 2 (212564_at), score: -0.63 KIAA0562KIAA0562 (204075_s_at), score: 0.66 KIAA1305KIAA1305 (220911_s_at), score: -0.71 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: 0.78 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: 0.68 LARSleucyl-tRNA synthetase (217810_x_at), score: 0.75 LEPREL2leprecan-like 2 (204854_at), score: -0.69 LIG1ligase I, DNA, ATP-dependent (202726_at), score: -0.67 LIN7Blin-7 homolog B (C. elegans) (219760_at), score: 0.64 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: 0.67 LMF2lipase maturation factor 2 (212682_s_at), score: -0.64 LMNB2lamin B2 (216952_s_at), score: -0.65 LOC100129500hypothetical protein LOC100129500 (212884_x_at), score: 0.65 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: 0.64 LOC391132similar to hCG2041276 (216177_at), score: 0.79 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: 0.65 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: -0.84 LRP2low density lipoprotein-related protein 2 (205710_at), score: -0.71 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: 0.75 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.85 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: -0.82 MAP7D1MAP7 domain containing 1 (217943_s_at), score: -0.64 MAPK7mitogen-activated protein kinase 7 (35617_at), score: -0.64 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: -0.73 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: -0.68 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: -0.64 MED15mediator complex subunit 15 (222175_s_at), score: -0.63 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: 0.74 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: -0.64 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: -0.65 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: 0.71 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: 0.65 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: -0.65 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: -0.7 MUC1mucin 1, cell surface associated (207847_s_at), score: -0.65 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: -0.74 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: 0.65 NARG1LNMDA receptor regulated 1-like (219378_at), score: 0.72 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: -0.68 NCALDneurocalcin delta (211685_s_at), score: 0.63 NCKAP1NCK-associated protein 1 (207738_s_at), score: 0.65 NDPNorrie disease (pseudoglioma) (206022_at), score: -0.9 NEFLneurofilament, light polypeptide (221805_at), score: 0.68 NEFMneurofilament, medium polypeptide (205113_at), score: 0.95 NF1neurofibromin 1 (211094_s_at), score: -0.9 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: -0.74 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: -0.73 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: 0.66 NPIPnuclear pore complex interacting protein (204538_x_at), score: -0.69 NUPL2nucleoporin like 2 (204003_s_at), score: 0.72 NXPH3neurexophilin 3 (221991_at), score: 0.73 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: -0.82 OLFML2Bolfactomedin-like 2B (213125_at), score: -1 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: 0.8 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: 0.72 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: 0.7 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: -0.68 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: -0.83 PARVBparvin, beta (37965_at), score: -0.82 PCDH9protocadherin 9 (219737_s_at), score: 0.94 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: -0.66 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: -0.69 PCNXpecanex homolog (Drosophila) (213159_at), score: -0.63 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: -0.69 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: -0.8 PDE7Bphosphodiesterase 7B (220343_at), score: 0.68 PHF7PHD finger protein 7 (215622_x_at), score: 0.75 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: -0.74 PISDphosphatidylserine decarboxylase (202392_s_at), score: -0.64 PITX1paired-like homeodomain 1 (208502_s_at), score: -0.68 PKN1protein kinase N1 (202161_at), score: -0.69 PLAUplasminogen activator, urokinase (211668_s_at), score: -0.74 PLCD1phospholipase C, delta 1 (205125_at), score: -0.64 PLK1polo-like kinase 1 (Drosophila) (202240_at), score: -0.69 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: -0.68 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: -0.69 POLSpolymerase (DNA directed) sigma (202466_at), score: -0.65 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.73 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: -0.68 PRKCDprotein kinase C, delta (202545_at), score: -0.8 PRMT3protein arginine methyltransferase 3 (213320_at), score: 0.86 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: -0.7 PXNpaxillin (211823_s_at), score: -0.91 RAB1ARAB1A, member RAS oncogene family (213440_at), score: 0.81 RAB22ARAB22A, member RAS oncogene family (218360_at), score: -0.64 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: 0.65 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: -0.65 RAG1AP1recombination activating gene 1 activating protein 1 (219125_s_at), score: 0.68 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: -0.67 RBM15BRNA binding motif protein 15B (202689_at), score: -0.62 RBM3RNA binding motif (RNP1, RRM) protein 3 (222026_at), score: 0.64 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: -0.86 RNF11ring finger protein 11 (208924_at), score: 0.66 RNF220ring finger protein 220 (219988_s_at), score: -0.85 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: 0.66 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.97 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: -0.66 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.86 RPS25ribosomal protein S25 (200091_s_at), score: 0.63 RPS6KA4ribosomal protein S6 kinase, 90kDa, polypeptide 4 (204632_at), score: -0.66 SAP30LSAP30-like (219129_s_at), score: -0.8 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: -0.73 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: -0.72 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.97 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: -0.7 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: 0.69 SEPHS1selenophosphate synthetase 1 (208939_at), score: -0.68 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: -0.76 SF1splicing factor 1 (208313_s_at), score: -0.87 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: -0.7 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: -0.64 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: -0.77 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: 0.73 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: 0.76 SLC39A14solute carrier family 39 (zinc transporter), member 14 (212110_at), score: 0.63 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: -0.77 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: 0.74 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.75 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: 0.64 SMAD3SMAD family member 3 (218284_at), score: -0.75 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: -0.73 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: -0.67 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.83 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: -0.71 ST20suppressor of tumorigenicity 20 (210242_x_at), score: 0.63 STIM1stromal interaction molecule 1 (202764_at), score: -0.78 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: 0.73 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: 0.79 TAF13TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa (205966_at), score: 0.71 TAPBPTAP binding protein (tapasin) (208829_at), score: -0.73 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: -0.69 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: 0.63 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: -0.68 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: -0.72 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.84 TLE4transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) (204872_at), score: 0.81 TMSB15Bthymosin beta 15B (214051_at), score: -0.64 TP53tumor protein p53 (201746_at), score: -0.73 TRAF5TNF receptor-associated factor 5 (204352_at), score: 0.68 TRIM21tripartite motif-containing 21 (204804_at), score: -0.75 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: 0.77 TSC2tuberous sclerosis 2 (215735_s_at), score: -0.67 TSKUtsukushin (218245_at), score: -0.73 TSPAN31tetraspanin 31 (203227_s_at), score: 0.65 TXLNAtaxilin alpha (212300_at), score: -0.67 UBTD1ubiquitin domain containing 1 (219172_at), score: -0.81 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: -0.69 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: 0.63 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.91 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: -0.77 WDR4WD repeat domain 4 (221632_s_at), score: 0.64 WDR42AWD repeat domain 42A (202249_s_at), score: -0.74 WDR6WD repeat domain 6 (217734_s_at), score: -0.89 XAB2XPA binding protein 2 (218110_at), score: -0.67 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: -0.85 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: 0.63 YTHDC2YTH domain containing 2 (213077_at), score: 0.68 ZC3H3zinc finger CCCH-type containing 3 (213445_at), score: -0.64 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: 0.7 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: -0.71 ZFAND1zinc finger, AN1-type domain 1 (218919_at), score: 0.63 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: 0.69 ZNF281zinc finger protein 281 (218401_s_at), score: 0.73 ZNF318zinc finger protein 318 (203521_s_at), score: -0.65 ZNF652zinc finger protein 652 (205594_at), score: 0.65
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |